Abstract
Diseases caused by Streptococcus suis are a significant economic and welfare concern in pigs as well as in humans. Several molecular methods have been applied to investigate S. suis strain diversity and identify phylogenetic groups. Multilocus sequence typing (MLST), commonly used to differentiate between S. suis strains, has been instrumental in identifying that the species is genetically highly diverse. Recent advances in whole-genome analysis have resulted in schemes permitting the classification of S. suis populations as pathogenic or non-pathogenic, or disease-associated or non-disease associated. Here, we review these and other molecular approaches that can be used for surveillance, outbreak tracking, preventative health management, effective treatment and control, as well as vaccine development, including PCR based-assays that are easy to apply in modest diagnostic settings and which allow for the rapid screening of a large number of isolates at relatively low cost, granting the identification of several major clonal complexes of the S. suis population.
1. Introduction
Streptococcus suis is a pathogen of pigs that can cause infections in humans who are in close contact with infected animals and/or contaminated pork-derived products, as well as those who have consumed raw pork products [1]. S. suis can cause meningitis, septicemia, endocarditis, and arthritis in humans [2,3,4]. Human S. suis cases have been reported in most countries of Western Europe, North and South America, Africa, Australia, New Zealand, India, Japan, and several other East and Southeast Asian countries, particularly China, Vietnam, and Thailand [1,3,4]. Indeed, S. suis has been shown to be responsible for thousands of human cases, of which 90.2% were in Asia, 8.5% in Europe, and 1.3% in other parts of the world [4].
Previously, S. suis had been classified into 35 serotypes (serotype 1/2, and 1–34) and which were then reduced to 33 serotypes because serotypes 32 and 34 were re-classified as S. orisratti [5,6,7,8,9]. In 2013, S. suis serotypes 20, 22, 26, and 33 were proposed to be removed from the S. suis taxon [10]: serotypes 20, 22, and 26 were proposed as Streptococcus parasuis [11,12], while serotype 33 was classified as Streptococcus ruminantium [13]. Hence, currently there are 29 true S. suis serotypes. Of them, serotype 2 is the most prevalent in both human and pig infections although cases caused by serotypes 4, 5, 9, 14, 16, 21, 24, and 31 have also been reported [1,2,3,4,14,15,16,17].
Studying the population structure and the genetic diversity of S. suis is helpful to understand the epidemiology of this organism as well as reveal clones or clonal groups with an apparently increased capacity to cause disease, or which are potentially associated with particular clinical manifestations. Several molecular typing techniques have been applied to the study of S. suis genetic diversity (Table 1); for example, pulse-field gel electrophoresis (PFGE) [18,19], randomly amplified polymorphic DNA (RAPD) [20,21], amplified fragment length polymorphism (AFLP) [22], polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [23], ribotyping [24,25], multilocus sequence typing (MLST) [26], and multilocus variable number tandem repeat analysis (MLVA) [27].

Table 1.
Characteristics of the molecular epidemiological methods for the Streptococcus suis study.
Of these techniques, MLST is a popular method that has been used throughout the world to determine the sequence types (STs) as well as clonal complexes (CCs) of S. suis strains isolated from pigs and humans, which has enhanced our understanding of the global S. suis epidemiology. More recently, the use of whole-genome sequencing (WGS; Table 1) has been instrumental in defining S. suis isolates as being members of epidemic/highly virulent (E/HV), virulent (V), or intermediately/weakly virulent (I/WV) groups [28]. Bayesian Analysis of Population Structure group (BAPS) [29] and minimum core genome group (MCG) analyses have also been used to characterize S. suis isolates [30,31]. Recently a pathotyping tool has been proposed to differentiate between disease-associated and non-disease associated groups [32]. Here, we review these current methods which have been applied to determine the population structure of S. suis isolates.
2. Multilocus Sequence Typing (MLST)
MLST is considered the gold standard to determine the structure of S. suis populations. This powerful method uses genetic variation that accumulates very slowly in housekeeping genes to investigate the genetic diversity of S. suis and its use has allowed global and long-term epidemiology. King et al. established the MLST scheme for S. suis in 2002, using seven different house-keeping genes (cpn60, dpr, recA, aroA, thrA, gki, and mutS) [26]. Since its establishment, this MLST scheme has been adopted by multiple laboratories throughout the world and used to determine the STs of S. suis strains isolated from pig and human cases of infection [1].
Thus far (28 November 2019), a total of 1245 STs have been recorded in the S. suis MLST database (https://pubmlst.org/ssuis/). This method has revealed the presence of many clonal complexes (CCs) within the S. suis population in the database. Among the different CCs identified, the most important causes of infections in human and pig have been CC1, CC16, CC20, CC25, CC28, CC94, CC104, CC233/379, and CC221/234 (Figure 1) [1,4,17,33]. Different CCs were found to be distributed in different regions of the world [1]. CC1 was found mostly in Europe, Asia (Cambodia, China, Korea, Japan, Thailand, and Vietnam), Australia, and South America (Argentina), while CC20 (ST20) was described as being important in the Netherlands [1,33,34,35,36,37]. Furthermore, ST7 (CC1), responsible for the 1998 and 2005 epidemics, was mostly present in China [1] and CC16 and CC94 were predominant in Europe, although human cases were reported in Thailand [17,33]. CC25 or CC28 were reported in North America and were also recovered in Thailand, Korea, Japan, and Australia [1,4,33,34,35,36,37]. Finally, CC104 (ST101, ST104, ST391-ST393, ST512-ST514), and CC233/379 (ST233 and ST379) were endemic to Thailand [1,33]. In North America, the structure of S. suis strains is more complicated and pathotypes are different from Europe and Asia [38]. Serotypes 1, 1/2, 2, 7, 14, and 23 as well as ST1, ST13, ST28, ST94, ST108, ST961, and ST977 have recently been described as pathogenic strains in the USA [38].
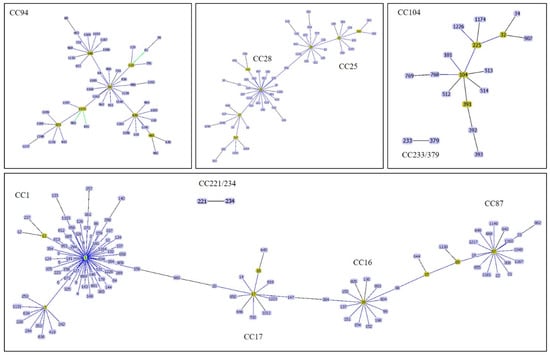
Figure 1.
A goeBURST analysis of major clonal complexes of S. suis causes of infections in human and pig. CC1 is related to CC17 whereas CC17 is closely related to CC16. CC16 and CC87 are related via ST15 and ST89. CC25 and CC28 are related via ST856. Other CCs are independent.
An alternative MLST approach using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS-MLST) has been reported to be more rapid in providing typing results than dideoxy sequencing [36]. While both MS-MLST and conventional MLST had 100% concordance in their classification of sequence types (STs), in addition to faster time-to-results, MS-MLST had lower labor requirements and per-isolate costs. MS-MLST analysis was easier to instrument when large numbers of isolates were involved [36]. However, MS-MLST requires a high initial investment in the MALDI-TOF MS equipment and software. Therefore, this approach may not be readily available to most laboratories, particularly those in developing countries.
3. Prediction of CC Using Polymerase Chain Reaction
Although informative, MLST is a high-cost and time-consuming method that is not ideal for screening large numbers of isolates. MLST is also unavailable in many laboratories in developing countries. Therefore, polymerase chain reaction (PCR)-based approaches [38,39,40,41,42] to identify the most important S. suis CCs (PCR-CC) were developed allowing for the rapid screening of a large number of isolates at a relatively modest cost (summarized in Table 2).

Table 2.
PCR-predicted important clonal complexes of S. suis isolates.
A first study reported that a variant of the serum opacity factor (ofs) can serve as a genetic marker to determine CCs of S. suis [39]. The report showed that a type-1 ofs was strongly associated with CC1, while type-3 ofs and type-4 ofs were associated with CC27. Importantly, CC25, CC28, and CC104 were included in CC27 with the less-stringent group definition (5 alleles sharing) used in that study [39]. However, since type-3 ofs was present in both CC94 and CC27, these two CCs could not be resolved. Similarly, CC25 and CC104 could not be differentiated from CC27.
A second study described a PCR assay to determine how pilus-associated gene profiles correlated with CCs of S. suis [40]. The results showed that such gene profiling could be used as a screening method for potentially hazardous S. suis groups. For example, genotype A of the pilus-associated gene profile was strongly associated with CC1, genotype B was associated with CC25 and CC27, and genotype C was related to CC104. Despite its enhanced discriminatory power, this assay cannot distinguish between CC25 and CC27, and multiple PCR reactions are necessary to identify the gene profiles for each genotype.
A third study reported a multiplex PCR that can be used to identify S. suis CCs that are relevant to human infections in a single reaction [41]. Of the 7 sequences encoded—2 hypothetical proteins (hp1 and hp2), 1 ribonuclease G (ribG), 1 peptidase (pep), 1 muramidase-like protein (mp), 1 zonula occludens toxin (zot), and 1 collagen adhesion (col)—only hp1, mp, pep, and col were selected as targets for PCR with the srtBCD primers described by Takamatsu et al. (2009). The test developed in that study differentiated between CC1, CC25, CC28, CC104, CC221/234, and CC233/379. However, the assay could not detect CC16, CC20, and CC94, and the report did not evaluate assays on CCs mostly related to pig infections.
A fourth report by Maneerat et al. (2013) demonstrated that RAPD using OPB-07, OPB-10, and OPB-17 could distinguish CC1, CC104, CC233/379, and ST336 [42]. This study revealed a high agreement of RAPD patterns among the three primers used with the specific pattern of each CC in each primer. Nonetheless, this study did not evaluate other S. suis CCs, such as CC25, CC28, and CC16.
A last report used random amplification of polymorphic DNA (RAPD) and 16S-23S rDNA intergenic spacer polymerase chain reaction-restriction fragment length polymorphism (16S-23S PCR-RFLP) to determine clonal complexes (CCs) of 684 S. suis isolates from pigs (n=72) and humans (n=612) [43]. The study also revealed that RAPD was better at distinguishing the major S. suis CCs than the PCR-RFLP method: the assay was capable of simultaneously distinguishing CC1, CC16, CC25, CC28, CC104, CC221/234, and CC233/379. RAPD with the OPB-10 primer clearly distinguished each CC in the study, and no cross-patterns were found. Finally, the study also showed that neither the OPB-06 nor OPB-11 primer could distinguish CC104 from CC233/379 isolates. The report showed that PCR-RFLP and DNA sequencing using the 16S-23S rDNA intergenic spacer could not clearly differentiate among most CCs [43]. Both techniques could classify S. suis isolates into four clusters: cluster 1 consisting of CC25, CC28, CC104, and CC233/379; cluster 2 consisting of CC221/234; cluster 3 consisting of CC16 (ST16); and cluster 4 consisting of CC1.
It is important to note that pig S. suis isolates are likely to be much more diverse than zoonotic human isolates. However, none of abovementioned studies evaluated S. suis CCs related to pig infections such as CC7, CC11, CC13/149, CC43/52, CC53/54, CC87, CC94, CC123, CC147, CC226/227, CC239/241, and CC20.
In addition to these PCR-based techniques, MLVA is a technique with high discriminatory power that has also been used to investigate S. suis. While PFGE could not differentiate between isolates, one MLVA scheme using nine loci (TR1-TR9) was successfully used to link and to sub-type S. suis ST7 isolates from two outbreaks that occurred in China in 1998 and 2005 [27]. Additional advantages of MLVA are that it is relatively easy to perform, rapid and reliable, and that it permits high-throughput screenings. MLVA also allows the comparison of results between laboratories and is useful in case of outbreak investigations.
4. Whole-Genome Sequencing Approaches
WGS approaches have increasingly been used to investigate S. suis isolates, including molecular determination of serotype [44], characterization of outbreaks [45,46], evaluation of S. suis reinfection [47], and to determine the population structure of S. suis isolates of serotype 2 belonging to ST25 and ST28 [48,49], as well as of serotype 9 [50]. WGS-based bacterial typing strategies commonly use one of two approaches: SNP (sequence)-based, or MLST (allele)-based. The SNP approach compares single nucleotide differences between isolates in comparison to a reference genome and is particularly useful to determine the clonal relationship between highly similar isolates. The MLST approach is an extension of conventional 7-gene MLST that expands the range of genes to the genome scale and can be roughly divided into core-genome-based MLST (cgMLST) and whole-genome-based MLST (wgMLST). Allele-based comparisons can be conducted using analysis tools available on the internet, such as BacWGSTdb (http://bacdb.org/BacWGSTdb/) [51], pubMLST (https://pubmlst.org/ssuis/), or the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/MLST/) [52].
The definition of virulence in S. suis remains controversial, and WGS-based approaches have played an important role in the efforts to identify virulence markers permitting to differentiate between commensal organisms and those prone to cause disease. A comparative genome hybridization (CGH) was used to analyze 55 S. suis isolates from different serotypes, recovered from different hosts, different clinical sources, and different geographical locations [53]. Clustering of CGH data divided S. suis isolates into 2 clusters. Cluster A exclusively contained virulent CC1 isolates of serotypes 1 and 2 isolates [53]. Cluster B, on the other hand, contained mainly a more divergent and heterogeneous group of serotype 7 and 9 isolates [53]. Another study used CGH on 39 isolates of different serotypes, sources, geographic locations, isolation years, and STs. This study revealed that the S. suis strains tested could be classified into three groups of differing levels of virulence: (i) epidemic and highly virulent (E/HV group), which included ST1, ST6, ST7, and ST11 isolates; (ii) virulent (V group), containing ST81, ST13, ST56, ST87, ST308, ST54, and ST53 isolates; and (iii) intermediately or weakly virulent (I/WV group), composed of isolates belonging to several STs that were all recovered from non-human sources [28].
The genomes of 375 S. suis isolates with detailed clinical phenotypes from pigs and humans from the United Kingdom and Vietnam were analyzed using a Bayesian Analysis of Population Structure (BAPS) [54]. The study showed clear genetic differences between systemic, respiratory, and non-clinical (carriage) S. suis isolates. Interestingly, systemic isolates had a smaller genome than respiratory and carriage isolates, and they tended to have an over-representation of virulence factors and a larger number of genes involved in defense functions [54]. Willemse et al. (2016) used BAPS to investigate the whole genomes of 98 S. suis isolates from human patients and pigs with invasive disease in the Netherlands, and 18 genomic complete and available S. suis sequences. The BAPS-based population grouping did not correlate with serotype but correlated well with the CC of the isolates. BAPS Group 1 comprised all CC13 isolates, BAPS Group 2 comprised most of the CC16 isolates, BAPS Group 4 comprised all CC1 isolates, BAPS Group 5 comprised both the CC27 and the CC29 isolates, and BAPS Group 6 comprised most CC20 isolates [29].
Minimum core genome sequence typing (MCG) was proposed by Chen et al. in 2013 [30]. These authors sequenced the genomes of 85 S. suis isolates belonging to 75 STs and defined an MCG of 876 genes and 58,501 SNPs. Using this variation, they reported seven MCG groups among their strain population. MCG Group 1 included all the highly virulent isolates of ST1 and the epidemic isolates of ST7, as well as all isolates from human infections and outbreaks, while the intermediate virulent STs, such as ST25 and ST28, were located in MCG Group 4 [30]. As MCG typing using whole-genome sequencing is unavailable and not cost effective in limited-resource countries, a single nucleotide polymorphisms-based typing method using PCR amplification and sequencing of six genes covering the distribution of 10 SNPs has been developed to identify the seven MCG groups [31]. This approach can categorize isolates into the correct MCG groups and offers a practical, rapid, and cost-effective assay for differentiating the population structure of S. suis.
Very recently, a genome-wide association study that took into consideration the observed clinical phenotype (invasive and asymptomatic) and used the discriminant analysis of principal components (DAPC model) implemented in the R package adegenet was conducted, which identified genetic markers of invasive disease and asymptomatic tonsillar carriage [32]. The identified markers were used to develop a pathotyping tool able to predict the virulence potential of S. suis isolates as disease-associated and non-disease associated [32] and also led to the development of a multiplex PCR assay for the identification of three S. suis genetic-markers that differentiated 115 S. suis isolates into disease-associated and non-disease-associated groups. The multiplex PCR assay revealed a good correlation compared to the observed clinical phenotypes, serotyping, virulence-associated gene (epf, mrp, and sly) profile, MLST, and MCG sequence type [32].
High-throughput sequencing systems are now available at many public health and hospital diagnostic laboratories, and the use of these systems in rapid diagnosis of S. suis has shown great potential. A report from Canada described that WGS (Illumina MiSeq) was instrumental in identifying S. suis serotype 2 ST25 as the causative agent of a human infection in a farmer, which had previously been ascribed to the viridans group Streptococcus by biochemical methods [55]. More recently, Hayashi et al. (2019) used Illumina short-reads to investigate two human S. suis cases in Japan. The genomic approach revealed that the isolates belonged to serotype 2 ST28, were derived from the same genetic lineage, and were closely related to S. suis strains previously isolated from pigs in that country [56]. The Oxford Nanopore MinION sequencer has also been used to rapidly generate a full-length S. suis consensus genome sequence, achieving a 99.4% average accuracy. Using optimized methods, this technology also permitted to determine the antibiotic resistance profile and sequence type of the isolates, with additional error corrections [57]. This report could build towards a standardization of the WGS workflow, for routine and low-cost disease diagnostics that are easy to perform, as well as have a short turnaround time.
5. Conclusions
Several methods exist to investigate strain diversity and identify phylogenetic groups of S. suis. Serotyping and MLST are still commonly used to differentiate S. suis strains. Advances in sequencing technologies, such as WGS, have allowed the determination of the population structure of S. suis strains and identified groups differing in virulence potential using Bayesian analysis, minimum core genome analysis, or genome-wide association. Collectively, PCR assays, which are easy to apply in routine diagnostic testing, permit the identification of several major groups of the S. suis population. Molecular tools could be used for surveillance and preventative health management through the detection of clones or strains that have genetic potential to cause disease in healthy animals. Furthermore, these techniques could be applied to track isolates in an outbreak, select strains for a vaccine, and develop effective treatment and control plans.
Author Contributions
Conceptualization: R.H., N.F., M.G., A.K. Formal analysis: R.H., A.K. Original draft preparation: R.H., A.K. Review and editing: N.F., M.G., A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Faculty of Science and Engineering, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus and the Kasetsart University Research and Development Institute (KURDI).
Acknowledgments
We thank the Faculty of Science and Engineering, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus and the Kasetsart University Research and Development Institute (KURDI), Bangkok, Thailand, for English editing assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes. Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Hlebowicz, M.; Jakubowski, P.; Smiatacz, T. Streptococcus suis Meningitis: Epidemiology, Clinical Presentation and Treatment. Vector Borne Zoonotic Dis. 2019, 19, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Huong, V.T.L.; Ha, N.; Huy, N.T.; Horby, P.; Nghia, H.D.T.; Thiem, V.D.; Zhu, X.; Hoa, N.T.; Hien, T.T.; Zamora, J.; et al. Epidemiology, Clinical manifestations, and outcomes of Streptococcus suis infection in Humans. Emerg. Infect. Dis. 2014, 20, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Sroka, J.; Zając, V.; Wasiński, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part I—Epidemiology. Ann. Agric. Environ. Med. 2017, 24, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Mittal, K.R.; Henrichsen, J. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 1989, 27, 2633–2636. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 1991, 29, 2590–2594. [Google Scholar] [CrossRef]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Isolation and characterization of Streptococcus suis capsular types 9–22. J. Vet. Diagn. Investig. 1991, 3, 60–65. [Google Scholar] [CrossRef]
- Higgins, R.; Gottschalk, M.; Boudreau, M.; Lebrun, A.; Henrichsen, J. Description of Six New Capsular Types (29–34) of Streptococcus Suis. J. Vet. Diagn. Investig. 1995, 7, 405–406. [Google Scholar] [CrossRef]
- Hill, J.E.; Gottschalk, M.; Brousseau, R.; Harel, J.; Hemmingsen, S.M.; Goh, S.H. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 2005, 107, 63–69. [Google Scholar] [CrossRef]
- Tien, L.H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA–DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef]
- Nomoto, R.; Maruyama, F.; Ishida, S.; Tohya, M.; Sekizaki, T.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. Int. J. Syst. Evol. Microbiol. 2015, 65, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current Taxonomical Situation of Streptococcus suis. Pathogens 2016, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Tohya, M.; Arai, S.; Tomida, J.; Watanabe, T.; Kawamura, Y.; Katsumi, M.; Ushimizu, M.; Ishida-Kuroki, K.; Yoshizumi, M.; Uzawa, Y.; et al. Defining the taxonomic status of Streptococcus suis serotype 33: The proposal for Streptococcus ruminantium sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3660–3665. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Oishi, K.; Akeda, Y. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect. Dis. 2015, 15, 392. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Oishi, K.; Sripakdee, S.; Boonkerd, N.; Polwichai, P.; Nakamura, S.; Uchida, R.; Sawanpanyalert, P.; Dejsirilert, S. Clonal dissemination of Streptococcus suis serotype 14 in Thailand. J. Med. Microbiol. 2009, 58, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Sawanpanyalert, P.; Boonnark, A.; Noithachang, W.; Sriyakum, D.; Simkum, S.; Chokngam, S.; Gottschalk, M.; Akeda, Y.; et al. Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 2011, 378, 960. Available online: https://doi.org/10.1016/S0140-6736(11)60923-9 (accessed on 27 January 2020). [CrossRef]
- Kerdsin, A.; Hatrongjit, R.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Akeda, Y.; Oishi, K. Emergence of Streptococcus suis serotype 9 infection in humans. J. Microbiol. Immunol. Infect. 2017, 50, 545–546. [Google Scholar] [CrossRef]
- Berthelot-Hérault, F.; Marois, C.; Gottschalk, M.; Kobisch, M. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J. Clin. Microbiol 2002, 40, 615–619. [Google Scholar] [CrossRef]
- Vela, A.I.; Goyache, J.; Tarradas, C.; Luque, I.; Mateos, A.; Moreno, M.A.; Borge, C.; Perea, J.A.; Domínguez, L.; Fernandez-Garayzabal, J.F. Analysis of Genetic Diversity of Streptococcus suis Clinical Isolates from Pigs in Spain by Pulsed-Field Gel Electrophoresis. J. Clin. Microbiol. 2003, 41, 2498–2502. [Google Scholar] [CrossRef] [PubMed]
- Chatellier, S.; Gottschalk, M.; Higgins, R.; Brousseau, R.; Harel, J. Relatedness of Streptococcus suisSerotype 2 Isolates from Different Geographic Origins as Evaluated by Molecular Fingerprinting and Phenotyping. J. Clin. Microbiol. 1999, 37, 362–366. [Google Scholar] [CrossRef]
- Cloutier, G.; D’Allaire, S.; Martinez, G.; Surprenant, C.; Lacouture, S.; Gottschalk, M. Epidemiology of Streptococcus suis serotype 5 infection in a pig herd with and without clinical disease. Vet. Microbiol. 2003, 97, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Rehm, T.; Baums, C.G.; Strommenger, B.; Beyerbach, M.; Valentin-Weigand, P.; Goethe, R. Amplified fragment length polymorphism of Streptococcus suis strains correlates with their profile of virulence-associated genes and clinical background. J. Med. Microbiol. 2007, 56, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Marois, C.; Le Devendec, L.; Gottschalk, M.; Kobisch, M. Molecular characterization of Streptococcus suis strains by 16S–23S intergenic spacer polymerase chain reaction and restriction fragment length polymorphism analysis. Can. J. Vet. Res. 2006, 70, 94–104. [Google Scholar]
- Harel, J.; Higgins, R.; Gottschalk, M.; Bigras-Poulin, M. Genomic relatedness among reference strains of different Streptococcus suis serotypes. Can. J. Vet. Res. 1994, 58, 259–262. [Google Scholar]
- Okwumabua, O.; Staats, J.; Chengappa, M.M. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping). J. Clin. Microbiol. 1995, 33, 968–972. [Google Scholar] [CrossRef]
- King, S.J.; Leigh, J.A.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Development of a Multilocus Sequence Typing Scheme for the Pig Pathogen Streptococcus suis: Identification of Virulent Clones and Potential Capsular Serotype Exchange. J. Clin. Microbiol. 2002, 40, 3671–3680. [Google Scholar] [CrossRef]
- Li, W.; Ye, C.; Jing, H.; Cui, Z.; Bai, X.; Jin, D.; Zheng, H.; Zhao, A.; Xu, Y.; Gottschalk, M.; et al. Streptococcus suis outbreak investigation using multiple-locus variable tandem repeat number analysis. Microbiol. Immunol. 2010, 54, 380–388. [Google Scholar] [CrossRef]
- Zheng, H.; Lan, R.; Zheng, X.; Cui, Z.; Liu, Z.; Bai, X.; Ji, S.; Gottschalk, M.; Xu, J. Comparative Genomic Hybridization Identifies Virulence Differences in Streptococcus suis. PLoS ONE 2014, 9, e87866. [Google Scholar] [CrossRef]
- Willemse, N.; Howell, K.J.; Weinert, L.A.; Heuvelink, A.; Pannekoek, Y.; Wagenaar, J.A.; Smith, H.E.; Van Der Ende, A.; Schultsz, C. An emerging zoonotic clone in the Netherlands provides clues to virulence and zoonotic potential of Streptococcus suis. Sci. Rep. 2016, 6, 28984. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, W.; Zheng, H.; Lan, R.; Wang, H.; Du, P.; Bai, X.; Ji, S.; Meng, Q.; Jin, N.; et al. Minimum core genome sequence typing of bacterial pathogens: A unified approach for clinical and public health microbiology. J. Clin. Microbiol. 2013, 51, 2582–2591. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, S.; Lan, R.; Liu, Z.; Bai, X.; Zhang, W.; Gottschalk, M.; Xu, J. Population Analysis of Streptococcus suis Isolates from Slaughtered Swine by Use of Minimum Core Genome Sequence Typing. J. Clin. Microbiol. 2014, 52, 3568–3572. [Google Scholar] [CrossRef] [PubMed]
- Wileman, T.M.; Weinert, L.A.; Howell, K.J.; Wang, J.; Peters, S.E.; Williamson, S.M.; Wells, J.M.; Langford, P.R.; Rycroft, A.N.; Wren, B.W.; et al. Pathotyping the Zoonotic Pathogen Streptococcus suis: Novel Genetic Markers to Differentiate Invasive Disease-Associated Isolates from Non-Disease-Associated Isolates from England and Wales. J. Clin. Microbiol. 2019, 57, e01712-18. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Akeda, Y.; Takeuchi, D.; Dejsirilert, S.; Gottschalk, M.; Oishi, K. Genotypic diversity of Streptococcus suis strains isolated from humans in Thailand. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-I.; Jeon, A.B.; Jung, B.-Y.; Byun, J.-W.; Gottschalk, M.; Kim, A.; Kim, J.W.; Kim, H.-Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017, 79, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Schultsz, C.; Jansen, E.; Keijzers, W.; Rothkamp, A.; Duim, B.; Wagenaar, J.A.; Van Der Ende, A. Differences in the Population Structure of Invasive Streptococcus suis Strains Isolated from Pigs and from Humans in the Netherlands. PLoS ONE 2012, 7, e33854. [Google Scholar] [CrossRef] [PubMed]
- Groves, M.D.; Jordan, D.; Chapman, T.A.; Al Jassim, R. Multilocus sequence typing of Australian Streptococcus suis type 2 by MALDI-TOF mass spectrometry analysis of PCR amplicons. Vet. Microbiol. 2015, 177, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Tark, D.; Moon, S.-H.; Han, J.-I.; Kim, W.-I.; Cho, H.-S. Sepsis Caused by Streptococcus suis Serotype 2 in a Eurasian River Otter (Lutra lutra) in the Republic of Korea. J. Wildl. Dis. 2018, 54, 866–869. [Google Scholar] [CrossRef]
- Estrada, A.A.; Gottschalk, M.; Rossow, S.; Rendahl, A.; Gebhart, C.; Marthaler, D.G. Serotype and Genotype (Multilocus Sequence Type) of Streptococcus suis Isolates from the United States Serve as Predictors of Pathotype. J. Clin. Microbiol. 2019, 57, e00377-19. [Google Scholar] [CrossRef]
- Takamatsu, D.; Osaki, M.; Tharavichitkul, P.; Takai, S.; Sekizaki, T. Allelic variation and prevalence of serum opacity factor among the Streptococcus suis population. J. Med. Microbiol. 2008, 57, 488–494. [Google Scholar] [CrossRef]
- Takamatsu, D.; Nishino, H.; Ishiji, T.; Ishii, J.; Osaki, M.; Fittipaldi, N.; Gottschalk, M.; Tharavichitkul, P.; Takai, S.; Sekizaki, T. Genetic organization and preferential distribution of putative pilus gene clusters in Streptococcus suis. Vet. Microbiol. 2009, 138, 132–139. [Google Scholar] [CrossRef]
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Hamada, S.; Oishi, K.; Akeda, Y. Development of a multiplex PCR assay to detect the major clonal complexes of Streptococcus suis relevant to human infection. J. Med. Microbiol. 2016, 65, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Maneerat, K.; Yongkiettrakul, S.; Kramomtong, I.; Tongtawe, P.; Tapchaisri, P.; Luangsuk, P.; Chaicumpa, W.; Gottschalk, M.; Srimanote, P. Virulence Genes and Genetic Diversity ofStreptococcus suisSerotype 2 Isolates from Thailand. Transbound. Emerg. Dis. 2013, 60, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Kidchana, A.; Meekhanon, N.; Hatrongjit, R.; Gottschalk, M.; Kerdsin, A. Application of random amplified polymorphism DNA and 16S-23S rDNA intergenic spacer polymerase chain reaction-restriction fragment length polymorphism to predict major Streptococcus suis clonal complexes isolated from humans and pigs. Mol. Cell. Probes 2019, 43, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Athey, T.B.T.; Teatero, S.; Lacouture, S.; Takamatsu, D.; Gottschalk, M.; Fittipaldi, N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016, 16, 162. [Google Scholar] [CrossRef]
- Huang, W.; Wang, M.; Hao, H.; Yang, R.; Xie, J.; Su, J.; Lin, M.; Cui, Y.; Jiang, Y. Genomic epidemiological investigation of a Streptococcus suis outbreak in Guangxi, China, 2016. Infect. Genet. Evol. 2019, 68, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zheng, H.; Zhou, J.; Lan, R.; Ye, C.; Jing, H.; Jin, D.; Cui, Z.; Bai, X.; Liang, J.; et al. Detection of Multiple Parallel Transmission Outbreak of Streptococcus suis Human Infection by Use of Genome Epidemiology, China, 2005. Emerg. Infect Dis. 2017, 23, 204–211. [Google Scholar] [CrossRef]
- Willemse, N.; van der Ende, A.; Schultsz, C. Reinfection with Streptococcus suis analysed by whole genome sequencing. Zoonoses Public Health 2019, 66, 179–183. [Google Scholar] [CrossRef]
- Athey, T.B.; Teatero, S.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Population Structure and Antimicrobial Resistance Profiles of Streptococcus suis Serotype 2 Sequence Type 25 Strains. PLoS ONE 2016, 11, e0150908. [Google Scholar] [CrossRef]
- Athey, T.B.T.; Auger, J.-P.; Teatero, S.; Dumesnil, A.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Complex Population Structure and Virulence Differences among Serotype 2 Streptococcus suis Strains Belonging to Sequence Type 28. PLoS ONE 2015, 10, e0137760. [Google Scholar] [CrossRef]
- Zheng, H.; Du, P.; Qiu, X.; Kerdsin, A.; Roy, D.; Bai, X.; Xu, J.; Vela, A.I.; Gottschalk, M. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 2018, 49, 1. [Google Scholar] [CrossRef]
- Ruan, Z.; Feng, Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016, 44, D682–D687. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- De Greeff, A.; Wisselink, H.J.; De Bree, F.M.; Schultsz, C.; Baums, C.G.; Thi, H.N.; Stockhofe-Zurwieden, N.; Smith, H.E. Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC Microbiol. 2011, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Weinert, L.A.; Chaudhuri, R.R.; Wang, J.; Peters, S.E.; Corander, J.; Jombart, T.; Baig, A.; Howell, K.J.; Vehkala, M.; Välimäki, N.; et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015, 6, 6740. Available online: https://doi.org/10.1038/ncomms7740 (accessed on 27 January 2020). [CrossRef] [PubMed]
- Gomez-Torres, J.; Nimir, A.; Cluett, J.; Aggarwal, A.; Elsayed, S.; Soares, D.; Teatero, S.; Chen, Y.; Gottschalk, M.; Fittipaldi, N. Human Case of Streptococcus suis Disease, Ontario, Canada. Emerg. Infect. Dis. 2017, 23, 2107–2109. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Tsukagoshi, H.; Sekizuka, T.; Ishikawa, D.; Imai, M.; Fujita, M.; Kuroda, M.; Saruki, N. Next-generation DNA sequencing analysis of two Streptococcus suis ST28 isolates associated with human infective endocarditis and meningitis in Gunma, Japan: A case report. Infect. Dis. 2018, 51, 62–66. [Google Scholar] [CrossRef]
- Tan, S.; Dvorak, C.M.T.; Estrada, A.A.; Gebhart, C.; Marthaler, D.G.; Murtaugh, M.P. MinION sequencing of Streptococcus suis allows for functional characterization of bacteria by multilocus sequence typing and antimicrobial resistance profiling. J. Microbiol. Methods 2019, 169, 105817. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).