Molecular and Phylogenetic Analysis of Tick-Borne Pathogens in Ticks Parasitizing Native Korean Goats (Capra hircus coreanae) in South Korea
Abstract
1. Introduction
2. Results
2.1. Identification of Ticks
2.2. Identification of TBPs
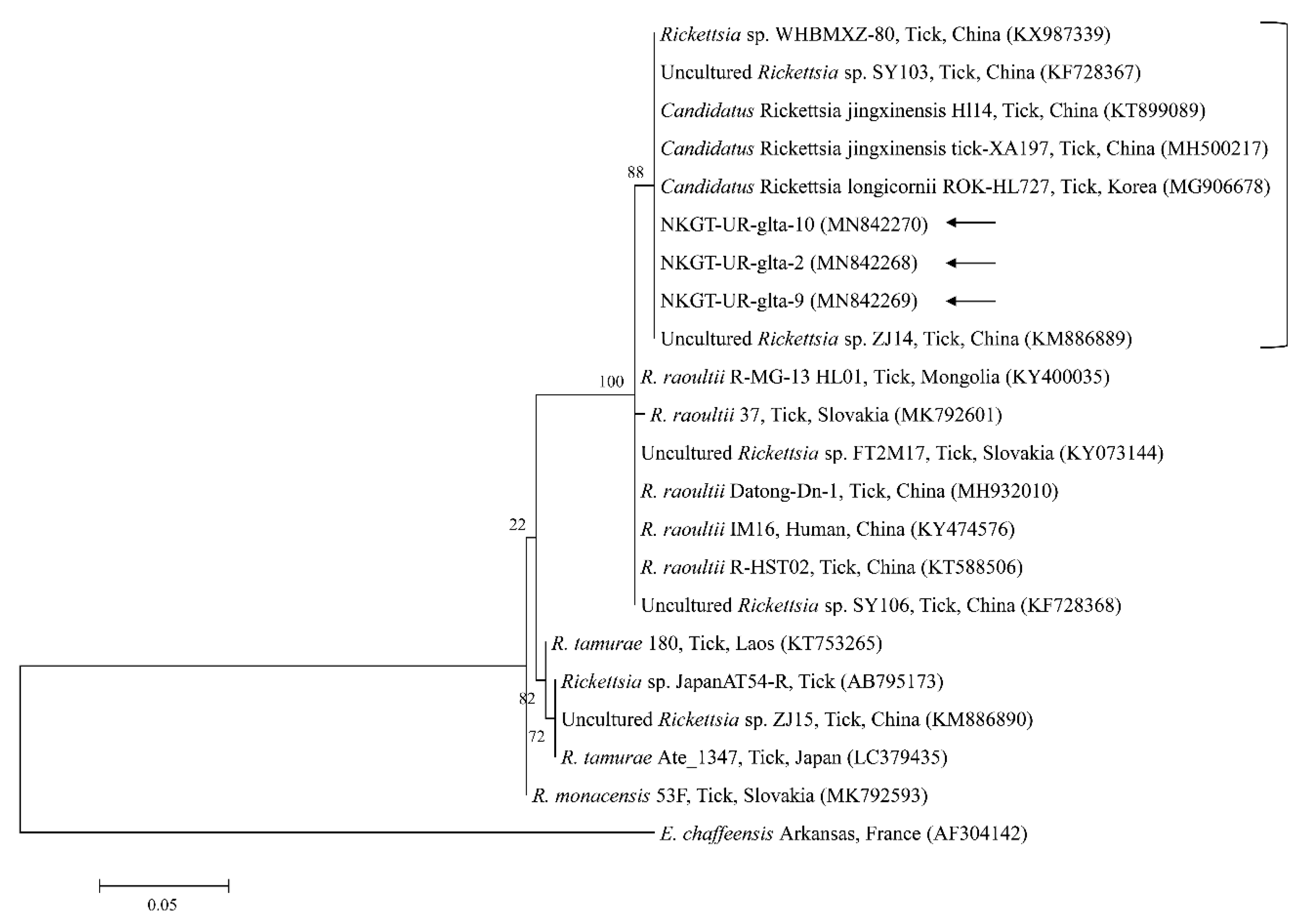
2.3. Molecular and Phylogenetic Analyses
3. Discussion
4. Materials and Methods
4.1. Tick Collection and Species Identification
4.2. Molecular Detection of Ticks and TBPs
4.3. DNA Cloning
4.4. Nucleotide Sequencing and Phylogeny
Supplementary Material
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Colwell, D.D.; Dantas-Torres, F.; Otranto, D. Vector-borne parasitic zoonoses: Emerging scenarios and new perspectives. Vet. Parasitol. 2011, 182, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G. Tick-borne infections of animals and humans: A common ground. Int. J. Parasitol. 2014, 44, 591–596. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Estrada-Pena, A.; Venzal, J.M.; Kocan, K.M.; Sonenshine, D.E. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. A J. Virtual Libr. 2008, 13, 6938–6946. [Google Scholar] [CrossRef] [PubMed]
- Salkeld, D.J.; Lane, R.S. Community ecology and disease risk: Lizards, squirrels, and the Lyme disease spirochete in California, USA. Ecology 2010, 91, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Moon, B.C.; Bae, B.K.; Shin, E.H.; Ko, Y.H.; Kim, Y.J.; Park, Y.H.; Chae, J.S. Genetic identification and phylogenetic analysis of Anaplasma and Ehrlichia species in Haemaphysalis longicornis collected from Jeju island, Korea. J. Bacteriol. Virol. 2009, 39, 257–267. [Google Scholar] [CrossRef]
- Kang, J.G.; Ko, S.; Kim, H.C.; Chong, S.T.; Klein, T.A.; Chae, J.B.; Jo, Y.S.; Choi, K.S.; Yu, D.H.; Park, B.K.; et al. Prevalence of Anaplasma and Bartonella spp. in ticks collected from Korean water deer (Hydropotes inermis argyropus). Korean J. Parasitol. 2016, 54, 87–91. [Google Scholar] [CrossRef]
- Kang, J.G.; Ko, S.; Smith, W.B.; Kim, H.C.; Lee, I.Y.; Chae, J.S. Prevalence of Anaplasma; Bartonella and Borrelia Species in Haemaphysalis longicornis collected from goats in North Korea. J. Vet. Sci. 2016, 17, 207–216. [Google Scholar] [CrossRef]
- Seo, M.G.; Lee, S.H.; VanBik, D.; Ouh, I.O.; Yun, S.H.; Choi, E.; Park, Y.S.; Lee, S.E.; Kim, J.W.; Cho, G.J.; et al. Detection and genotyping of Coxiella burnetii and Coxiella-like bacteria in horses in South Korea. PLoS ONE 2016, 11, e0156710. [Google Scholar] [CrossRef]
- Seo, M.G.; Ouh, I.O.; Lee, H.; Geraldino, P.J.L.; Rhee, M.H.; Kwon, O.D.; Kwak, D. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet. Microbiol. 2018, 226, 15–22. [Google Scholar] [CrossRef]
- Moon, K.H.; Lee, S.; Choi, C.Y.; Kim, S.Y.; Kang, C.W.; Lee, K.K.; Yun, Y.M. Investigation of Theileria sp. from ticks and roe deer (Capreolus pygargus) in jeju island. J. Vet. Clin. 2014, 31, 6–10. [Google Scholar] [CrossRef]
- Noh, Y.; Lee, Y.S.; Kim, H.C.; Chong, S.T.; Klein, T.A.; Jiang, J.; Richards, A.L.; Lee, H.K.; Kim, S.Y. Molecular detection of Rickettsia species in ticks collected from the southwestern provinces of the Republic of Korea. Parasites Vectors 2017, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; An, H.; Lee, J.S.; O’Guinn, M.L.; Kim, H.C.; Chong, S.T.; Zhang, Y.; Song, D.; Burrus, R.G.; Bao, Y.; et al. Molecular characterization of Haemaphysalis longicornis-borne rickettsiae, Republic of Korea and China. Ticks Tick Borne Dis. 2018, 9, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.Y.; Seo, M.G.; Lee, S.H.; Byun, J.W.; Oem, J.K.; Kwak, D. Molecular and serologic detection of Coxiella burnetii in native Korean goats (Capra hircus coreanae). Vet. Microbiol. 2014, 173, 152–155. [Google Scholar] [CrossRef]
- Seong, G.; Han, Y.J.; Chae, J.B.; Chae, J.S.; Yu, D.H.; Lee, Y.S.; Park, J.; Park, B.K.; Yoo, J.G.; Choi, K.S. Detection of Anaplasma sp. in Korean native goats (Capra aegagrus hircus) on Jeju Island, Korea. Korean J. Parasitol. 2015, 53, 765–769. [Google Scholar] [CrossRef]
- Chae, J.B.; Cho, Y.S.; Cho, Y.K.; Kang, J.G.; Shin, N.S.; Chae, J.S. Epidemiological investigation of tick species from near domestic animal farms and cattle; goat; and wild boar in Korea. Korean J. Parasitol. 2019, 57, 319–324. [Google Scholar] [CrossRef]
- Chong, S.T.; Kim, H.C.; Lee, I.Y.; Kollars, T.M., Jr.; Sancho, A.R.; Sames, W.J.; Chae, J.S.; Klein, T.A. Seasonal distribution of ticks in four habitats near the demilitarized zone, Gyeonggi-do (Province), Republic of Korea. Korean J. Parasitol. 2013, 51, 319–325. [Google Scholar] [CrossRef]
- Battilani, M.; De Arcangeli, S.; Balboni, A.; Dondi, F. Genetic diversity and molecular epidemiology of Anaplasma. Infect. Genet. Evol. 2017, 49, 195–211. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, M.; Wang, Z.; Wang, J.; Peng, Y.; Li, Y.; Guan, G.; Luo, J.; Yin, H. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl. Environ. Microbiol. 2012, 78, 464–470. [Google Scholar] [CrossRef]
- Ooshiro, M.; Zakimi, S.; Matsukawa, Y.; Katagiri, Y.; Inokuma, H. Detection of Anaplasma bovis and Anaplasma phagocytophilum from cattle on Yonaguni Island, Okinawa, Japan. Vet. Parasitol. 2008, 154, 360–364. [Google Scholar] [CrossRef]
- Uilenberg, G. International collaborative research: Significance of tick-borne hemoparasitic diseases to world animal health. Vet. Parasitol. 1995, 57, 19–41. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Liu, Z.; Chen, Z.; Yang, J.; He, H.; Guan, G.; Liu, A.; Ren, Q.; Niu, Q.; et al. An epidemiological survey of Theileria infections in small ruminants in central China. Vet. Parasitol. 2014, 200, 198–202. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, K.T.; Kwon, O.D.; Ock, Y.; Kim, T.; Choi, D.; Kwak, D. Novel detection of Coxiella spp., Theileria luwenshuni, and T. ovis endosymbionts in deer keds (Lipoptena fortisetosa). PLoS ONE 2016, 11, e0156727. [Google Scholar] [CrossRef]
- Madariaga, M.G.; Rezai, K.; Trenholme, G.M.; Weinstein, R.A. Q fever: A biological weapon in your backyard. Lancet Infect. Dis. 2003, 3, 709–721. [Google Scholar] [CrossRef]
- Duron, O.; Noël, V.; McCoy, K.D.; Bonazzi, M.; Sidi-Boumedine, K.; Morel, O.; Vavre, F.; Zenner, L.; Jourdain, E.; Durand, P.; et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen; Coxiella burnetii. PLoS Pathog. 2015, 11, e1004892. [Google Scholar] [CrossRef]
- Merhej, V.; Angelakis, E.; Socolovschi, C.; Raoult, D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect. Genet. Evol. 2014, 25, 122–137. [Google Scholar] [CrossRef]
- Parola, P.; Paddock, C.D.; Raoult, D. Tick-borne rickettsioses around the world: Emerging diseases challenging old concepts. Clin. Microbiol. Rev. 2005, 18, 719–756. [Google Scholar] [CrossRef]
- Liu, H.; Li, Q.; Zhang, X.; Li, Z.; Wang, Z.; Song, M.; Wei, F.; Wang, S.; Liu, Q. Characterization of rickettsiae in ticks in northeastern China. Parasites Vectors. 2016, 9, 498. [Google Scholar] [CrossRef]
- Guo, W.P.; Wang, Y.H.; Lu, Q.; Xu, G.; Luo, Y.; Ni, X.; Zhou, E.M. Molecular detection of spotted fever group rickettsiae in hard ticks, northern China. Transbound. Emerg. Dis. 2019, 66, 1587–1596. [Google Scholar] [CrossRef]
- Ministry of Agriculture, Food and Rural Affairs, South Korea (MAFRA). Major Statistics of Agriculture, Food and Rural Affairs during 2019; Ministry of Agriculture, Food and Rural Affairs: Sejong, Korea, 2019.
- Barker, S.C.; Walker, A.R. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa 2014, 3816, 1–144. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Reis, C.; Cote, M.; Paul, R.E.; Bonnet, S. Questing ticks in suburban forest are infected by at least six tick-borne pathogens. Vector Borne Zoonotic Dis. 2011, 11, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Kim, S.J.; Kang, J.G.; Won, S.; Lee, H.; Shin, N.S.; Choi, K.S.; Youn, H.Y.; Chae, J.S. Molecular detection of Bartonella grahamii and B. schoenbuchensis-related species in Korean water deer (Hydropotes inermis argyropus). Vector Borne Zoonotic Dis. 2013, 13, 415–418. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Fukushi, S.; Tani, H.; Fukuma, A.; Taniguchi, S.; Toda, S.; Shimazu, Y.; Yano, K.; Morimitsu, T.; Ando, K.; et al. Sensitive and specific PCR systems for detection of both Chinese and Japanese severe fever with thrombocytopenia syndrome virus strains and prediction of patient survival based on viral load. J. Clin. Microbiol. 2014, 52, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Casati, S.; Sager, H.; Gern, L.; Piffaretti, J.C. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann. Agric. Environ. Med. 2006, 13, 65–70. [Google Scholar]
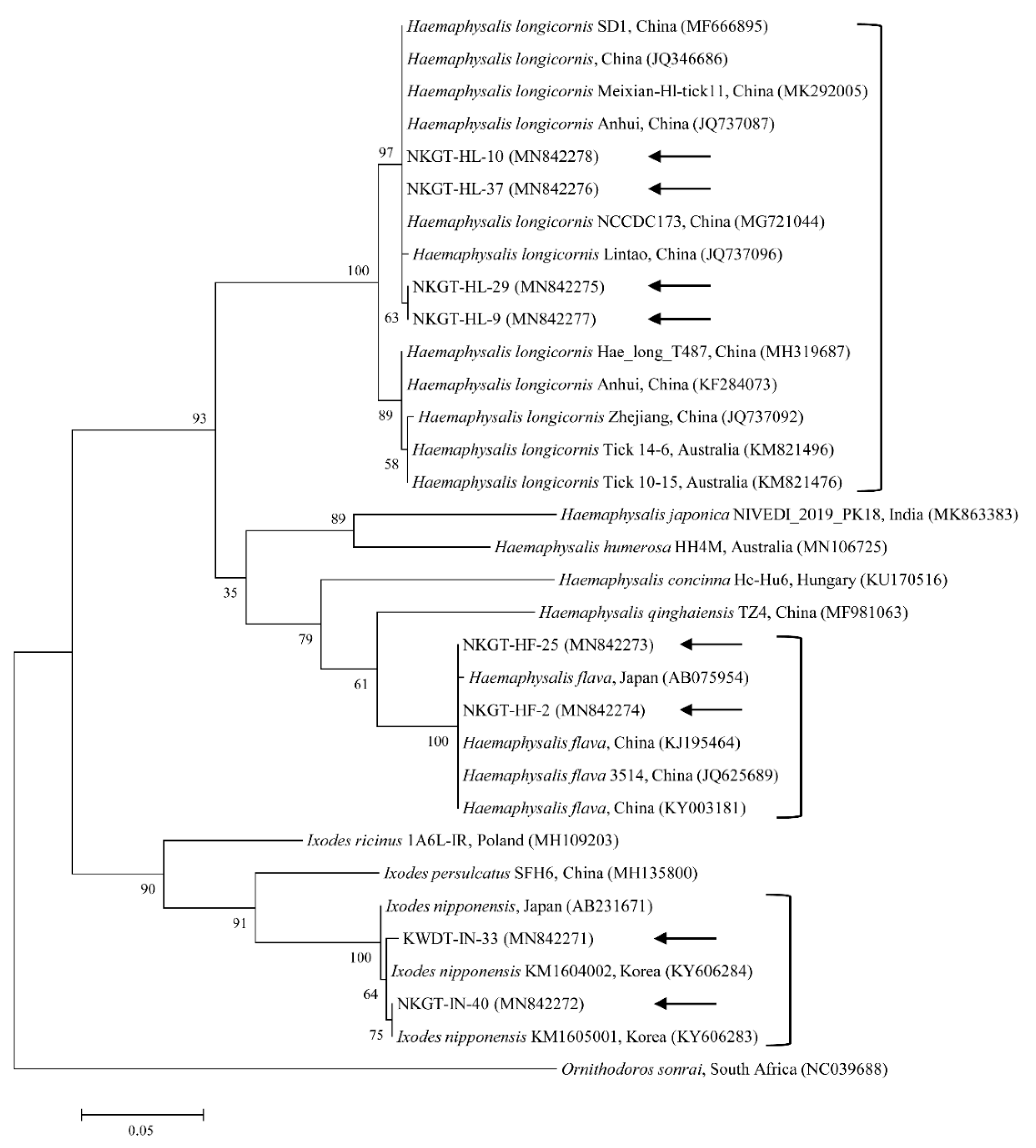
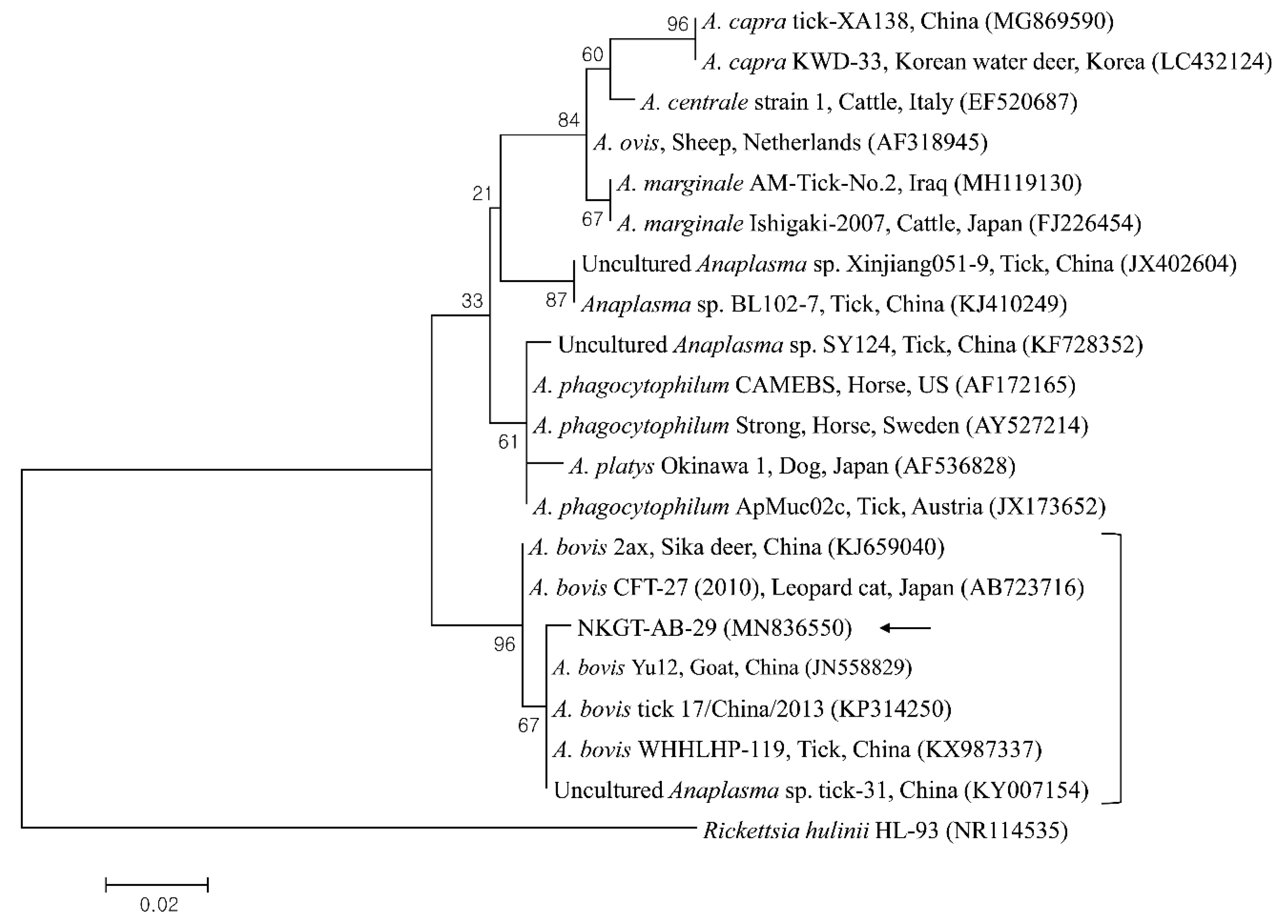
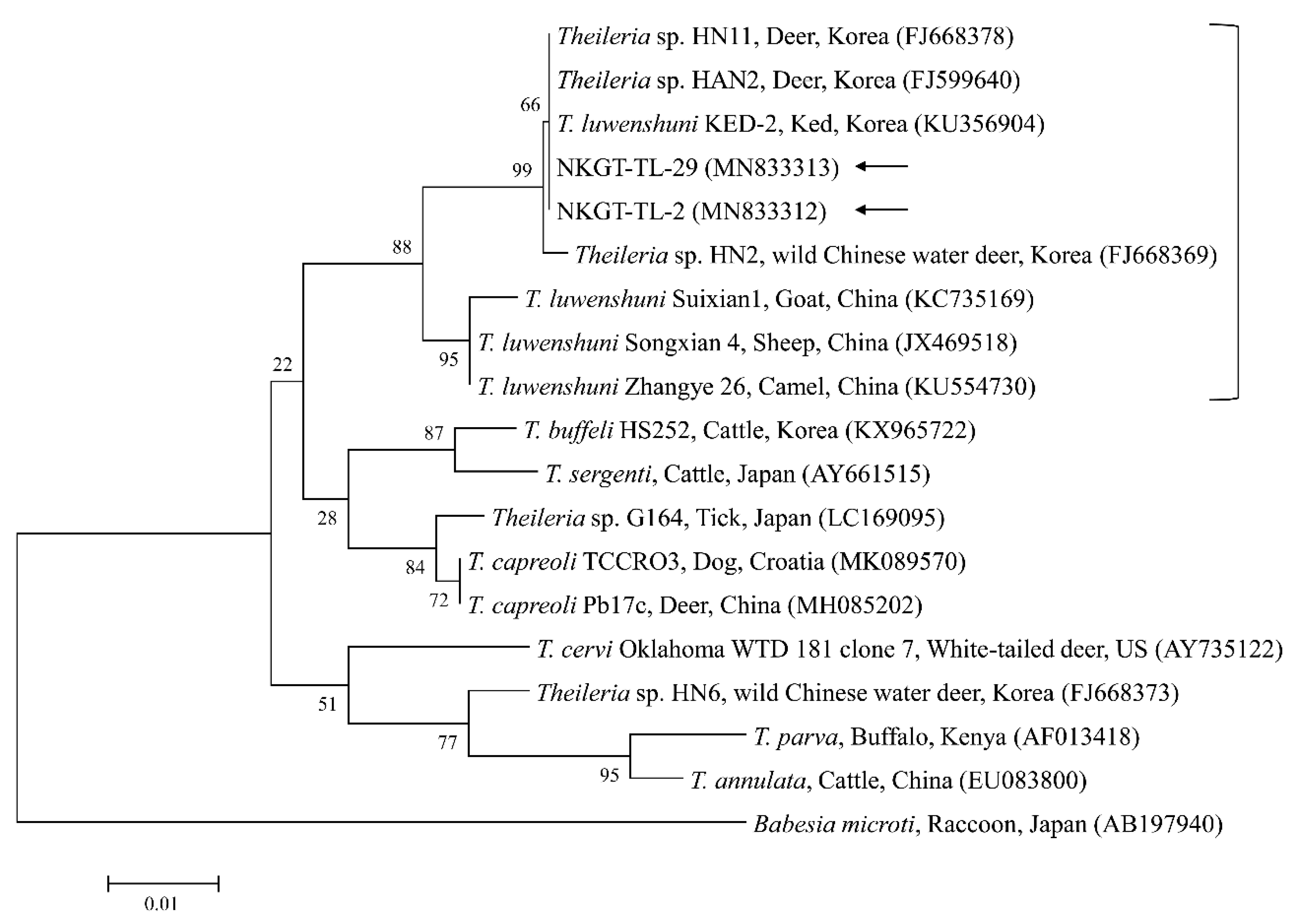
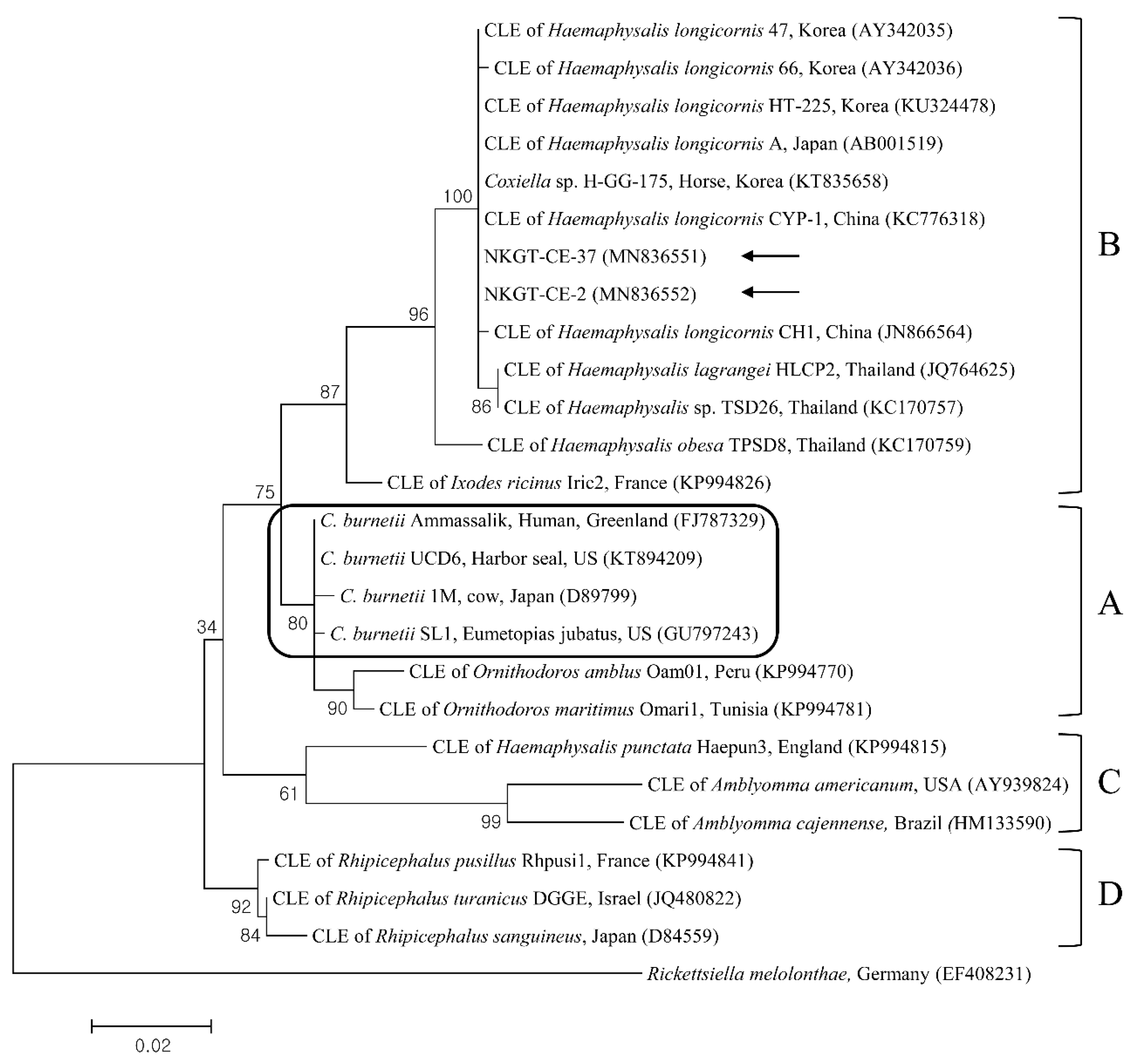

| Species | Stage | Tick Pool (n) | A. bovis (16S) | T. luwenshuni (18S) | Coxiella-like Endosymbiont (16S) | Candidatus R. Longicornii (16S) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive (%) | 95% CI | Positive (%) | 95% CI | Positive (%) | 95% CI | Positive (%) | 95% CI | |||
| Haemaphysalis longicornis | Nymph | 15 | 1 (6.7) | 0–19.3 | 1 (6.7) | 0–19.3 | 1 (6.7) | 0–19.3 | 8 (53.3) | 28.1–78.6 |
| Adult | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 (100) | 100 | |
| Haemaphysalis flava | Nymph | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Adult | 4 | 0 | 0 | 1 (25.0) | 0–67.4 | 1 (25.0) | 0–67.4 | 0 | 0 | |
| Ixodes nipponensis | Nymph | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Adult | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 40 | 1 (2.5) | 0–7.3 | 2 (5.0) | 0–11.8 | 2 (5.0) | 0–11.8 | 18 (45.0) | 29.6–60.4 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, M.-G.; Kwon, O.-D.; Kwak, D. Molecular and Phylogenetic Analysis of Tick-Borne Pathogens in Ticks Parasitizing Native Korean Goats (Capra hircus coreanae) in South Korea. Pathogens 2020, 9, 71. https://doi.org/10.3390/pathogens9020071
Seo M-G, Kwon O-D, Kwak D. Molecular and Phylogenetic Analysis of Tick-Borne Pathogens in Ticks Parasitizing Native Korean Goats (Capra hircus coreanae) in South Korea. Pathogens. 2020; 9(2):71. https://doi.org/10.3390/pathogens9020071
Chicago/Turabian StyleSeo, Min-Goo, Oh-Deog Kwon, and Dongmi Kwak. 2020. "Molecular and Phylogenetic Analysis of Tick-Borne Pathogens in Ticks Parasitizing Native Korean Goats (Capra hircus coreanae) in South Korea" Pathogens 9, no. 2: 71. https://doi.org/10.3390/pathogens9020071
APA StyleSeo, M.-G., Kwon, O.-D., & Kwak, D. (2020). Molecular and Phylogenetic Analysis of Tick-Borne Pathogens in Ticks Parasitizing Native Korean Goats (Capra hircus coreanae) in South Korea. Pathogens, 9(2), 71. https://doi.org/10.3390/pathogens9020071





