Appraisal of a Simple and Effective RT-qPCR Assay for Evaluating the Reverse Transcriptase Activity in Blood Samples from HIV-1 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
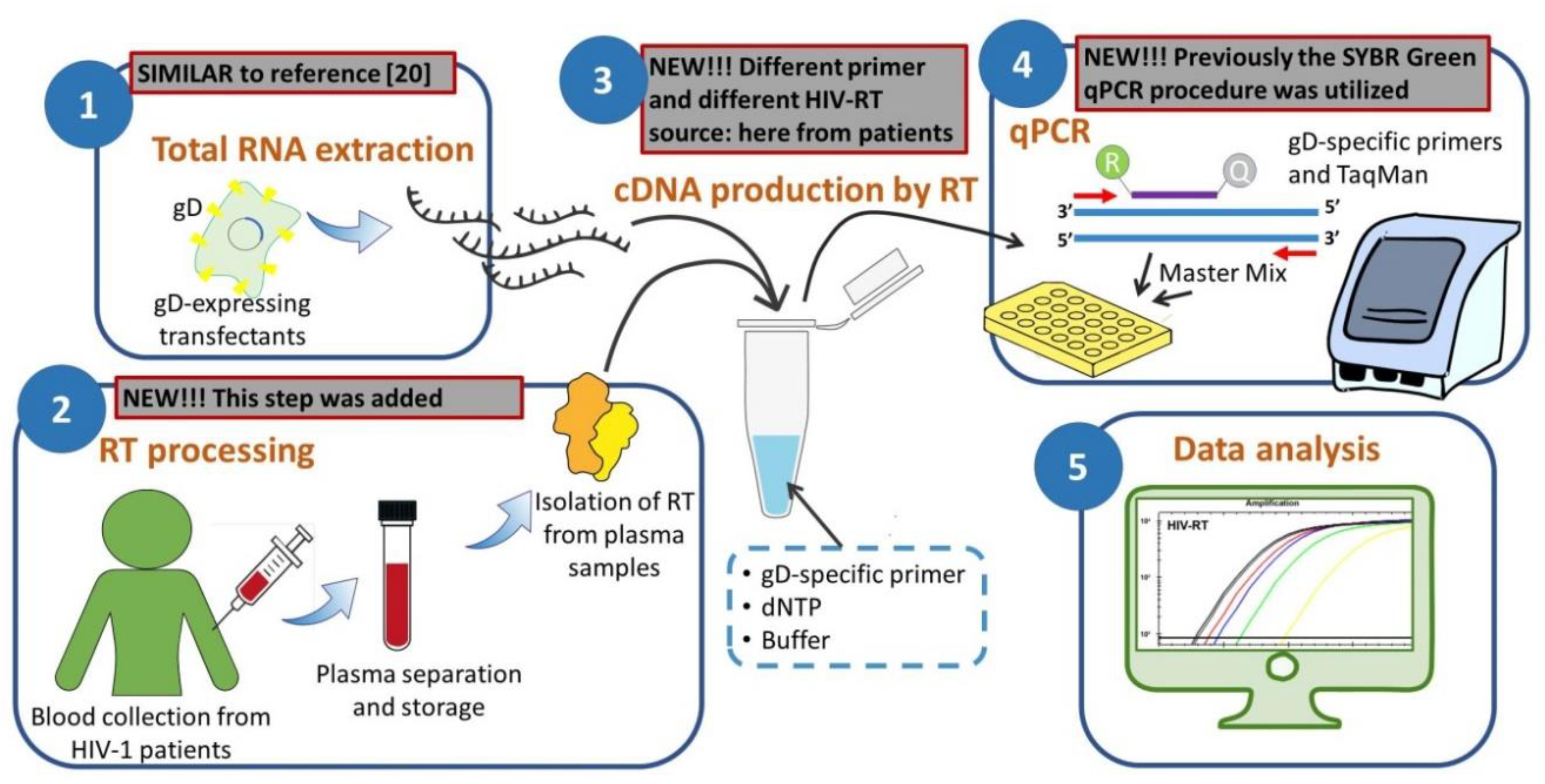
2.2. Isolation of HIV RT from Plasma Samples
2.3. Preparation of RNA Template by glycoprotein D (gD)-Enriched RNA Extraction
2.4. Primers and Probe for RT Activity Assay
2.5. RT Activity Assay
2.6. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rausch, J.W.; Le Grice, S.F.J. Characterizing the Latent HIV-1 Reservoir in Patients with Viremia Suppressed on cART: Progress, Challenges, and Opportunities. Curr. HIV Res. 2020, 18, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gurule, E.E.; Brennan, T.P.; Gerold, J.M.; Kwon, K.J.; Hosmane, N.N.; Kumar, M.R.; Beg, S.A.; Capoferri, A.A.; Ray, S.C.; et al. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. USA 2018, 115, E2575–E2584. [Google Scholar] [CrossRef] [PubMed]
- Sufka, S.A.; Ferrari, G.; Gryszowka, V.E.; Wrin, T.; Fiscus, S.A.; Tomaras, G.D.; Staats, H.F.; Patel, D.D.; Sempowski, G.D.; Hellmann, N.S.; et al. Prolonged CD4+ cell/virus load discordance during treatment with protease inhibitor-based highly active antiretroviral therapy: Immune response and viral control. J. Infect. Dis. 2003, 187, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Eberle, J.; Seibl, R. A new method for measuring reverse transcriptase activity by ELISA. J. Virol. Methods 1992, 40, 347–356. [Google Scholar] [CrossRef]
- Somogyi, P.A.; Gyuris, A.; Foldes, I. A solid phase reverse transcriptase micro-assay for the detection of human immunodeficiency virus and other retroviruses in cell culture supernatants. J. Virol. Methods 1990, 27, 269–276. [Google Scholar] [CrossRef]
- Boni, J.; Pyra, H.; Schupbach, J. Sensitive detection and quantification of particle-associated reverse transcriptase in plasma of HIV-1-infected individuals by the product-enhanced reverse transcriptase (PERT) assay. J. Med. Virol. 1996, 49, 23–28. [Google Scholar] [CrossRef]
- DeStefano, J.J.; Alves Ferreira-Bravo, I. A highly sensitive aptamer-based HIV reverse transcriptase detection assay. J. Virol. Methods 2018, 257, 22–28. [Google Scholar] [CrossRef]
- Greengrass, V.L.; Turnbull, S.P.; Hocking, J.; Dunne, A.L.; Tachedjian, G.; Corrigan, G.E.; Crowe, S.M. Evaluation of a low cost reverse transcriptase assay for plasma HIV-1 viral load monitoring. Curr. HIV Res. 2005, 3, 183–190. [Google Scholar] [CrossRef]
- Heneine, W.; Yamamoto, S.; Switzer, W.M.; Spira, T.J.; Folks, T.M. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 1995, 171, 1210–1216. [Google Scholar] [CrossRef]
- Lerma, J.; Palacin, J.A. The expanded suprapubic area as a skin donor site in the treatment of congenital absence of the vagina. Plast. Reconstr. Surg. 2000, 105, 2631–2632. [Google Scholar] [CrossRef]
- Lovatt, A.; Black, J.; Galbraith, D.; Doherty, I.; Moran, M.W.; Shepherd, A.J.; Griffen, A.; Bailey, A.; Wilson, N.; Smith, K.T. High throughput detection of retrovirus-associated reverse transcriptase using an improved fluorescent product enhanced reverse transcriptase assay and its comparison to conventional detection methods. J. Virol. Methods 1999, 82, 185–200. [Google Scholar] [CrossRef]
- Malmsten, A.; Shao, X.W.; Aperia, K.; Corrigan, G.E.; Sandstrom, E.; Kallander, C.F.; Leitner, T.; Gronowitz, J.S. HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J. Med. Virol. 2003, 71, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Malmsten, A.; Shao, X.W.; Sjodahl, S.; Fredriksson, E.L.; Pettersson, I.; Leitner, T.; Kallander, C.F.; Sandstrom, E.; Gronowitz, J.S. Improved HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J. Med. Virol. 2005, 76, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Pizzato, M.; Erlwein, O.; Bonsall, D.; Kaye, S.; Muir, D.; McClure, M.O. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods 2009, 156, 1–7. [Google Scholar] [CrossRef]
- Pyra, H.; Boni, J.; Schupbach, J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc. Natl. Acad. Sci. USA 1994, 91, 1544–1548. [Google Scholar] [CrossRef]
- Sears, J.F.; Khan, A.S. Single-tube fluorescent product-enhanced reverse transcriptase assay with Ampliwax (STF-PERT) for retrovirus quantitation. J. Virol. Methods 2003, 108, 139–142. [Google Scholar] [CrossRef]
- Silver, J.; Maudru, T.; Fujita, K.; Repaske, R. An RT-PCR assay for the enzyme activity of reverse transcriptase capable of detecting single virions. Nucleic Acids Res. 1993, 21, 3593–3594. [Google Scholar] [CrossRef]
- Vermeire, J.; Naessens, E.; Vanderstraeten, H.; Landi, A.; Iannucci, V.; Van Nuffel, A.; Taghon, T.; Pizzato, M.; Verhasselt, B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS ONE 2012, 7, e50859. [Google Scholar] [CrossRef]
- Frezza, C.; Balestrieri, E.; Marino-Merlo, F.; Mastino, A.; Macchi, B. A novel, cell-free PCR-based assay for evaluating the inhibitory activity of antiretroviral compounds against HIV reverse transcriptase. J. Med. Virol. 2014, 86, 1–7. [Google Scholar] [CrossRef]
- Marino-Merlo, F.; Frezza, C.; Papaianni, E.; Valletta, E.; Mastino, A.; Macchi, B. Development and evaluation of a simple and effective RT-qPCR inhibitory assay for detection of the efficacy of compounds towards HIV reverse transcriptase. Appl. Microbiol. Biotechnol. 2017, 101, 8249–8258. [Google Scholar] [CrossRef]
- Macchi, B.; Balestrieri, E.; Ascolani, A.; Hilburn, S.; Martin, F.; Mastino, A.; Taylor, G.P. Susceptibility of primary HTLV-1 isolates from patients with HTLV-1-associated myelopathy to reverse transcriptase inhibitors. Viruses 2011, 3, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Macchi, B.; Balestrieri, E.; Frezza, C.; Grelli, S.; Valletta, E.; Marcais, A.; Marino-Merlo, F.; Turpin, J.; Bangham, C.R.; Hermine, O.; et al. Quantification of HTLV-1 reverse transcriptase activity in ATL patients treated with zidovudine and interferon-alpha. Blood Adv. 2017, 1, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Medici, M.A.; Sciortino, M.T.; Perri, D.; Amici, C.; Avitabile, E.; Ciotti, M.; Balestrieri, E.; De Smaele, E.; Franzoso, G.; Mastino, A. Protection by herpes simplex virus glycoprotein D against Fas-mediated apoptosis: Role of nuclear factor kappaB. J. Biol. Chem. 2003, 278, 36059–36067. [Google Scholar] [CrossRef] [PubMed]
- Andreoni, M.; Sarmati, L.; Parisi, S.G.; Ercoli, L.; Rocchi, G. Efficient and reproducible new semimicromethod for the detection and titration of HIV in human plasma. J. Med. Virol. 1992, 38, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Coombs, R.W.; Collier, A.C.; Allain, J.P.; Nikora, B.; Leuther, M.; Gjerset, G.F.; Corey, L. Plasma viremia in human immunodeficiency virus infection. N. Engl. J. Med. 1989, 321, 1626–1631. [Google Scholar] [CrossRef]
- Lathey, J.L.; Fiscus, S.A.; Rasheed, S.; Kappes, J.C.; Griffith, B.P.; Elbeik, T.; Spector, S.A.; Reichelderfer, P.S. Optimization of quantitative culture assay for human immunodeficiency virus from plasma. Plasma Viremia Group Laboratories of the AIDS Clinical Trials Group (National Institute of Allergy and Infectious Diseases). J. Clin. Microbiol. 1994, 32, 3064–3067. [Google Scholar] [CrossRef]
- Rusert, P.; Fischer, M.; Joos, B.; Leemann, C.; Kuster, H.; Flepp, M.; Bonhoeffer, S.; Gunthard, H.F.; Trkola, A. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology 2004, 326, 113–129. [Google Scholar] [CrossRef]
- Cornall, A.; Sharma, L.; Solomon, A.; Gorry, P.R.; Crowe, S.M.; Cameron, P.U.; Lewin, S.R. A novel, rapid method to detect infectious HIV-1 from plasma of persons infected with HIV-1. J. Virol. Methods 2010, 165, 90–96. [Google Scholar] [CrossRef]
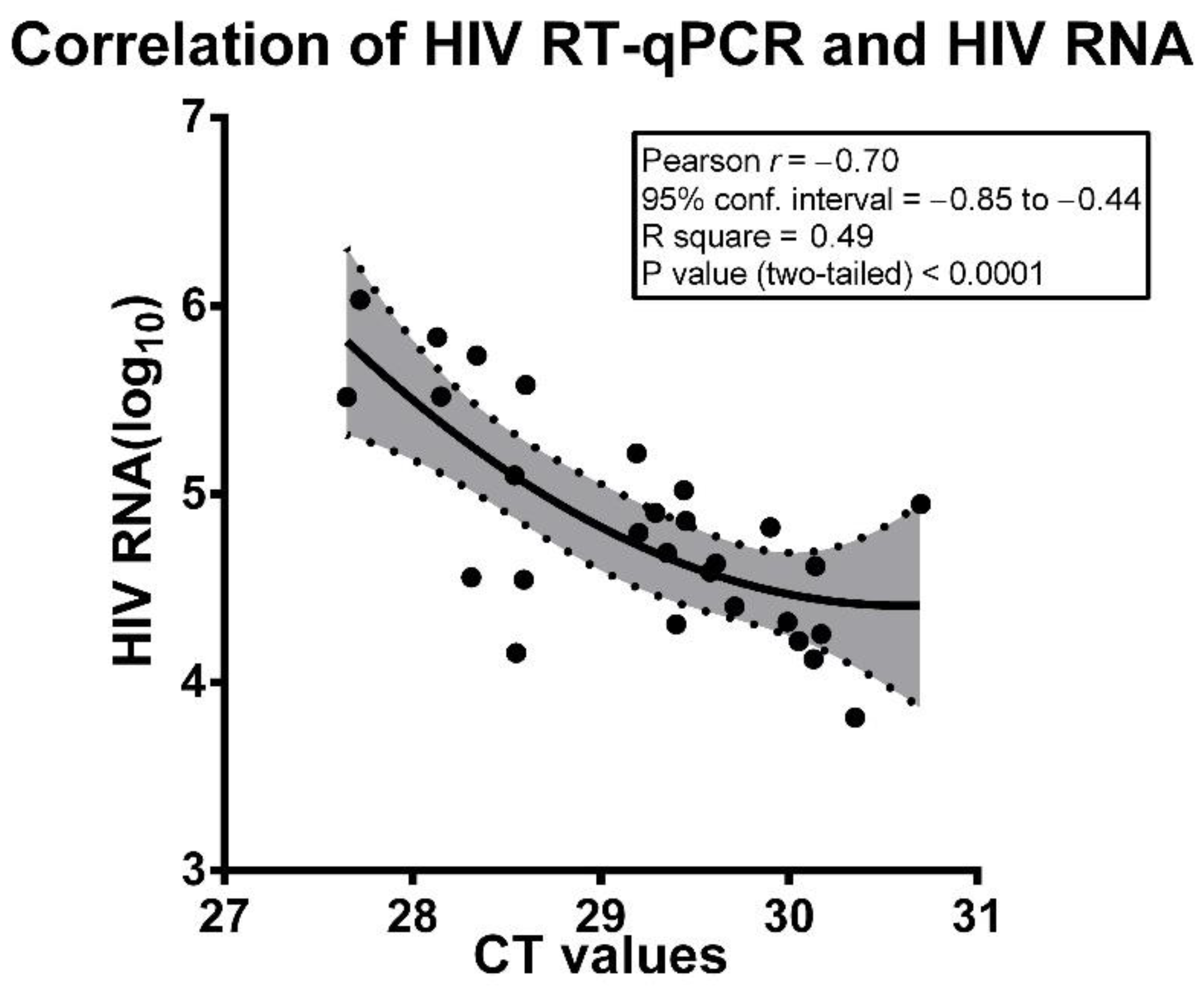
| Patient ID | HIV-1 RNA (Copies/mL) | HIV-1 RNA Copies/mL (log10) | Mean CT Values |
|---|---|---|---|
| 1 | 6523 | 3.81 | 30.35 |
| 2 | 13,304 | 4.12 | 30.13 |
| 3 | 14,377 | 4.16 | 28.55 |
| 4 | 16,558 | 4.22 | 30.05 |
| 5 | 18,053 | 4.26 | 30.17 |
| 6 | 20,400 | 4.31 | 29.40 |
| 7 | 20,931 | 4.32 | 29.99 |
| 8 | 25,300 | 4.40 | 29.71 |
| 9 | 35,157 | 4.55 | 28.59 |
| 10 | 36,196 | 4.56 | 28.31 |
| 11 | 38,664 | 4.59 | 29.58 |
| 12 | 41,521 | 4.62 | 30.14 |
| 13 | 42,770 | 4.63 | 29.61 |
| 14 | 48,786 | 4.69 | 29.35 |
| 15 | 62,329 | 4.79 | 29.20 |
| 16 | 66,681 | 4.82 | 29.90 |
| 17 | 72,111 | 4.86 | 29.45 |
| 18 | 79,849 | 4.90 | 29.29 |
| 19 | 88,801 | 4.95 | 30.70 |
| 20 | 104,832 | 5.02 | 29.44 |
| 21 | 126,006 | 5.10 | 28.54 |
| 22 | 165,295 | 5.22 | 29.19 |
| 23 | 328,191 | 5.52 | 27.65 |
| 24 | 330,013 | 5.52 | 28.15 |
| 25 | 379,959 | 5.58 | 28.60 |
| 26 | 544,912 | 5.74 | 28.34 |
| 27 | 683,104 | 5.83 | 28.13 |
| 28 | 1,075,865 | 6.03 | 27.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macchi, B.; Frezza, C.; Marino-Merlo, F.; Minutolo, A.; Stefanizzi, V.; Balestrieri, E.; Cerva, C.; Sarmati, L.; Andreoni, M.; Grelli, S.; et al. Appraisal of a Simple and Effective RT-qPCR Assay for Evaluating the Reverse Transcriptase Activity in Blood Samples from HIV-1 Patients. Pathogens 2020, 9, 1047. https://doi.org/10.3390/pathogens9121047
Macchi B, Frezza C, Marino-Merlo F, Minutolo A, Stefanizzi V, Balestrieri E, Cerva C, Sarmati L, Andreoni M, Grelli S, et al. Appraisal of a Simple and Effective RT-qPCR Assay for Evaluating the Reverse Transcriptase Activity in Blood Samples from HIV-1 Patients. Pathogens. 2020; 9(12):1047. https://doi.org/10.3390/pathogens9121047
Chicago/Turabian StyleMacchi, Beatrice, Caterina Frezza, Francesca Marino-Merlo, Antonella Minutolo, Valeria Stefanizzi, Emanuela Balestrieri, Carlotta Cerva, Loredana Sarmati, Massimo Andreoni, Sandro Grelli, and et al. 2020. "Appraisal of a Simple and Effective RT-qPCR Assay for Evaluating the Reverse Transcriptase Activity in Blood Samples from HIV-1 Patients" Pathogens 9, no. 12: 1047. https://doi.org/10.3390/pathogens9121047
APA StyleMacchi, B., Frezza, C., Marino-Merlo, F., Minutolo, A., Stefanizzi, V., Balestrieri, E., Cerva, C., Sarmati, L., Andreoni, M., Grelli, S., & Mastino, A. (2020). Appraisal of a Simple and Effective RT-qPCR Assay for Evaluating the Reverse Transcriptase Activity in Blood Samples from HIV-1 Patients. Pathogens, 9(12), 1047. https://doi.org/10.3390/pathogens9121047







