Genomic Analysis of Aeromonas veronii C198, a Novel Mcr-3.41-Harboring Isolate from a Patient with Septicemia in Thailand
Abstract
1. Introduction
2. Results and Discussion
2.1. General Characteristic of A. veronii C198
2.2. Genome Features
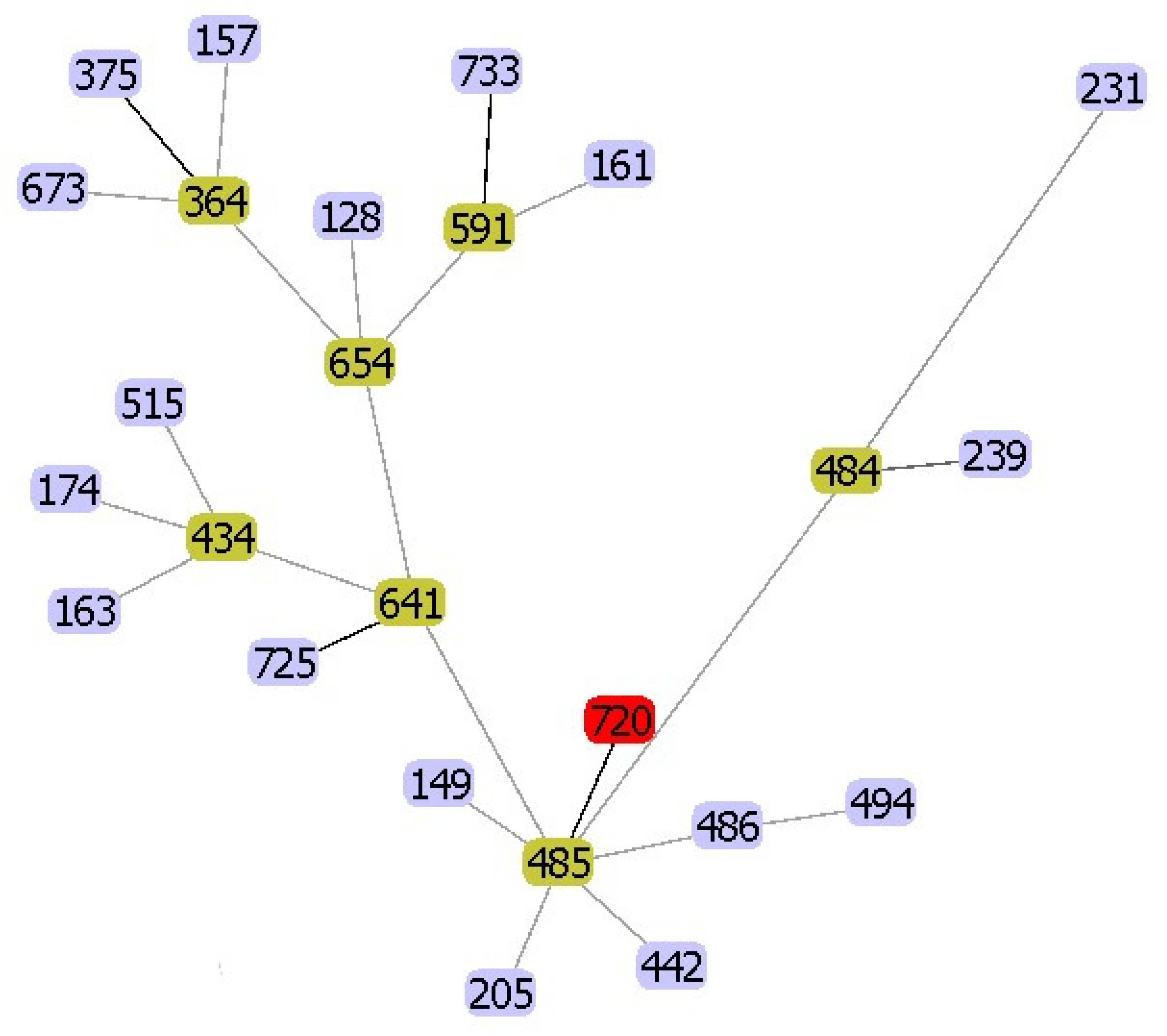
2.3. Phylogenetic Analysis
2.4. Antimicrobial Susceptibility and Resistance Genes
2.5. Virulence Factors
3. Materials and Methods
3.1. Bacterial Strain
3.2. Antimicrobial Susceptibility Testing
3.3. Whole-Genome Sequencing
3.4. Analysis of the Whole-Genome Sequence
3.5. Accession Numbers
3.6. Ethics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Tekedar, H.C.; Kumru, S.; Blom, J.; Perkins, A.D.; Griffin, M.J.; Abdelhamed, H.; Karsi, A.; Lawrence, M.L. Comparative genomics of Aeromonas veronii: Identification of a pathotype impacting aquaculture globally. PLoS ONE 2019, 14, e0221018. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.; Torri, A.; Amadori, E.; Rossi, A.; Bartolini, G.; Casadei, C.; Frassineti, G.L. Aeromonas veronii biovar veronii and sepsis-infrequent complication of biliary drainage placement: A case report. World J. Clin. Cases 2019, 7, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yu, L.; Nan, Z.; Zhang, P.; Kan, B.; Yan, D.; Su, J. Taxonomy, virulence genes and antimicrobial resistance of Aeromonas isolated from extra-intestinal and intestinal infections. BMC Infect Dis. 2019, 19, 158. [Google Scholar] [CrossRef]
- Wu, C.J.; Chen, P.L.; Hsueh, P.R.; Chang, M.C.; Tsai, P.J.; Shih, H.I.; Wang, H.C.; Chou, P.H.; Ko, W.C. Clinical implications of species identification in monomicrobial Aeromonas bacteremia. PLoS ONE 2015, 10, e0117821. [Google Scholar] [CrossRef]
- Nwaiwu, O.; Aduba, C.C. An in silico analysis of acquired antimicrobial resistance genes in Aeromonas plasmids. AIMS Microbiol. 2020, 6, 75–91. [Google Scholar]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: A review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, C.; Sun, Q.; Schwarz, S.; Ou, Y.; Yang, L.; Huang, Z.; Eichhorn, I.; Walsh, T.R.; Wang, Y.; et al. Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 2018, 62, e00404-18. [Google Scholar] [CrossRef]
- Lamy, B.; Kodjo, A.; Laurent, F. Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 2009, 47, 1234–1237. [Google Scholar] [CrossRef]
- Wu, C.J.; Chen, P.L.; Tang, H.J.; Chen, H.M.; Tseng, F.C.; Shih, H.I.; Hung, Y.P.; Chung, C.H.; Ko, W.C. Incidence of Aeromonas bacteremia in Southern Taiwan: Vibrio and Salmonella bacteremia as comparators. J. Microbiol. Immunol. Infect 2014, 47, 145–148. [Google Scholar] [CrossRef][Green Version]
- Turner, P.; Willemse, C.; Phakaudom, K.; Zin, T.W.; Nosten, F.; McGready, R. Aeromonas spp. bacteremia in pregnant women, Thailand-Myanmar border, 2011. Emerg. Infect Dis. 2012, 18, 1522–1523. [Google Scholar] [CrossRef] [PubMed]
- Chompoonuch, S.; Wangsomboonsiri, W.; Wongprasit, P.; Sungkanuparph, S.; Phakdeekitcharoen, B. Aeromonas hydrophila sepsis with septic embolism and rhabdomyolysis in a chronic iron overload haemodialysis patient treated with deferoxamine. NDT Plus 2009, 2, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Janma, J.; Linasmita, P.; Changsirikulchai, S. A report of peritonitis from Aeromonas sobria in a peritoneal dialysis (PD) patient with necrotizing fasciitis. J Med Assoc Thai 2015, 98 (Suppl. S10), 150–153. [Google Scholar]
- Ling, Z.; Yin, W.; Li, H.; Zhang, Q.; Wang, X.; Wang, Z.; Ke, Y.; Wang, Y.; Shen, J. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 2017, 61, e01272-17. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, W.; Wang, S.; Shen, Z.; Wang, Y.; Zhang, Q. A novel transposon, Tn6518, mediated transfer of mcr-3 variant in ESBL-producing Aeromonas veronii. Infect. Drug Resist. 2020, 13, 893–899. [Google Scholar] [CrossRef]
- Tekedar, H.C.; Arick, M.A., II; Hsu, C.Y.; Thrash, A.; Blom, J.; Lawrence, M.L.; Abdelhamed, H. Identification of antimicrobial resistance determinants in Aeromonas veronii strain MS-17-88 recovered from channel catfish (Ictalurus punctatus). Front Cell Infect Microbiol. 2020, 10, 348. [Google Scholar] [CrossRef]
- Gonçalves Pessoa, R.B.; de Oliveira, W.F.; Marques, D.; Dos Santos Correia, M.T.; de Carvalho, E.; Coelho, L. The genus Aeromonas: A general approach. Microb. Pathog. 2019, 130, 81–94. [Google Scholar] [CrossRef]
- Li, T.; Raza, S.; Yang, B.; Sun, Y.; Wang, G.; Sun, W.; Qian, A.; Wang, C.; Kang, Y.; Shan, X. Aeromonas veronii infection in commercial freshwater fish: A potential threat to public health. Animals 2020, 10, 608. [Google Scholar] [CrossRef]
- Clinical Laboratory Standard Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; CLSI Document M45-A2; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome. Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Vielva, L.; de Toro, M.; Lanza, V.F.; de la Cruz, F. PLACNETw: A web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 2017, 33, 3796–3798. [Google Scholar] [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | A. veronii C198 |
|---|---|
| Sequence Type | ST720 |
| Antimicrobial susceptibility (μg/mL) * | |
| Amoxicillin–clavulanic acid | 32/16 (R) |
| Ampicillin–sulbactam | 32/16 (R) |
| Piperacillin–tazobactam | 2/4 (S) |
| Cefepime | 0.25 (S) |
| Cefotaxime | 0.25 (S) |
| Cefoxitin | 1 (S) |
| Ceftazidime | 0.5 (S) |
| Ertapenem | 0.5 (S) |
| Imipenem | 0.125 (S) |
| Meropenem | 0.125 (S) |
| Amikacin | 4 (S) |
| Gentamicin | 1 (S) |
| Tetracycline | 32 (R) |
| Ciprofloxacin | 0.25 (S) |
| Levofloxacin | 0.25 (S) |
| Chloramphenicol | 2 (S) |
| Trimethoprim–sulfamethoxazole | 2/38 (S) |
| Colistin | 2 (ND **) |
| Antimicrobial and resistance genes | |
| β-lactam | blacphA3 |
| blaOXA-12 | |
| Tetracycline | tetA |
| Colistin | mcr-3 |
| Fluoroquinolone and tetracycline | adeF |
| Fluoroquinolone, diaminopyrimidine, and phenicol | rsmA |
| elfamycin | EF-Tu (R234F) # |
| Virulence genes | |
| Adherence | Flagella |
| Mannose-sensitive hemagglutinin | |
| Tap type IV pili | |
| Type I fimbria | |
| Secretion system | Type II secretion system |
| Type III secretion system | |
| Type VI secretion system | |
| Toxins | Aerolysin |
| Hemolysin III | |
| Hemolysin, HlyA | |
| Thermostable hemolysin | |
| Strain | Accession No. | Source | Country | MCR | cphA | OXA-12 | OXA-21 | TEM | SHV | AAC(6’) | AAC(3) | aadA | APH(3’)-Ia | APH(6)-Id | qnr | tet | mphA | floR | dfrA | sul | arr-3 | cat | adeF | rsmA | qacEdelta1 | EF-Tu Mutant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C198 | JACEGL000000000 | Human | Thailand | 3.41 | A3 | + | A | + | + | R234F | ||||||||||||||||
| 126-14 | PPTE00000000 | Human | China | 3.25 | A3 | + | D | + | + | R234F | ||||||||||||||||
| MGYG-HGUT-02529 | CABMOE000000000 | Human | China | 3.3 | A3 | + | + | IIa | A16 | A | A15 | 1 | B3 | + | + | + | R234F | |||||||||
| ZJ12-3 | UETM00000000 | Human | China | 3.3 | A3 | + | + | IIa | A16 | A | A15 | 1 | B3 | + | + | + | R234F | |||||||||
| AVNIH1 | CP014774 | Human | USA | A3 | + | 134 | Iic, Ib10 | IIg | A2 | B4 | D, E | + | + | A12 | 1, 2 | + | + | + | R234F | |||||||
| AVNIH2 | LRBO00000000 | Human | USA | A3 | + | E | + | + | R234F | |||||||||||||||||
| AER39 | AGWT00000000 | Human | USA | A3 | + | + | + | R234F | ||||||||||||||||||
| AMC35 | AGWW00000000 | Human | USA | A3 | + | + | + | R234F | ||||||||||||||||||
| BVH46 | NKWS01000000 | Human | USA | A3 | + | + | + | R234F | ||||||||||||||||||
| FDAARGOS_632 | CP044060 | Human | USA | A3 | + | + | + | R234F | ||||||||||||||||||
| AER397 | AGWV00000000 | Human | USA | A6 | + | + | + | R234F | ||||||||||||||||||
| CCM4359 | MRZR00000000 | Human | USA | A6 | + | + | + | |||||||||||||||||||
| CECT4257T | CDDK00000000 | Human | USA | A6 | + | + | + | |||||||||||||||||||
| FC951 | PKSR00000000 | Human | India | A3 | + | + | + | R234F | ||||||||||||||||||
| VBF557 | LXJN00000000 | Human | India | A3 | R234F | |||||||||||||||||||||
| 312M | RHDQ00000000 | Human | Brazil | A3 | + | + | + | R234F | ||||||||||||||||||
| BC88 | CAAKNH000000000 | Human | Australia | A7 | + | + | + | R234F | ||||||||||||||||||
| UDRT09 | JAAQQM000000000 | Fish | Thailand | A3 | + | 116 | S2 | E | + | + | R234F | |||||||||||||||
| CNRT12 | JAAQQN000000000 | Fish | Thailand | A3 | + | + | + | R234F | ||||||||||||||||||
| NK07 | JAAQQQ000000000 | Fish | Thailand | A3 | + | A2 | A | + | A12 | 1 | + | + | + | R234F | ||||||||||||
| TH0426 | CP012504 | Catfish | China | A3 | + | + | + | R234F | ||||||||||||||||||
| CB51 | CP015448 | Fish | China | A3 | + | + | R234F | |||||||||||||||||||
| JC529 | CP058912 | Fish | China | A3 | + | + | + | R234F | ||||||||||||||||||
| A8-AHP | CP046407 | Fish | India | A3 | + | + | + | + | R234F | |||||||||||||||||
| Ae52 | BDGY00000000 | Goldfish | Sri-Lanka | A3 | + | A2 | + | + | A, D | + | A12 | 1 | II | + | + | + | ||||||||||
| AG-5.28.6 | NNSE00000000 | Fish | Greece | A3 | + | + | + | |||||||||||||||||||
| PDB | NMUS00000000 | Fish | Greece | A3 | + | + | + | |||||||||||||||||||
| B48 | WSFS00000000 | Fish | Brazil | A3 | + | + | + | R234F | ||||||||||||||||||
| ML09-123 | PPUW00000000 | Catfish | USA | + | + | + | R234F | |||||||||||||||||||
| MS-17-88 | RAWX00000000 | Catfish | USA | 3, 7.1 | A3 | + | D | + | A3 | B7 | + | + | R234F | |||||||||||||
| TTU2014-141AME | LKKD00000000 | Cattle | USA | A3 | + | E | + | + | ||||||||||||||||||
| TTU2014-115ASC | LKJS00000000 | Cattle | USA | A3 | + | + | + | R234F | ||||||||||||||||||
| Z2-7 | UETI00000000 | Pork | China | 3.3 | A3 | + | D | A15 | 1 | + | + | + | R234F | |||||||||||||
| HX3 | CP040717 | Alligator | China | 3.3 | A3 | + | D | + | + | R234F | ||||||||||||||||
| 17ISAe | CP028133 | Discus fish | Korea | 3.6, 3.8 | A3 | + | Ib-cr | A | A1 | 1 | + | + | + | + | R234F | |||||||||||
| CCM7244 | MRZQ00000000 | Surface water | Germany | A6 | + | + | + | R234F | ||||||||||||||||||
| AK236 | NKXF00000000 | Lake water | France | A3 | + | + | + | |||||||||||||||||||
| ARB3 | JRBE00000000 | Pond water | Japan | A6 | + | + | + | |||||||||||||||||||
| KLG5 | CAAKNL000000000 | River | England | A3 | + | + | + | R234F | ||||||||||||||||||
| B565 | NC_015424 | Sediment | China | A6 | + | + | + | R234F | ||||||||||||||||||
| Pamvotica | MRUI00000000 | Sediment | Greece | A3 | + | + | + | R234F |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatrongjit, R.; Kerdsin, A.; Takeuchi, D.; Wongsurawat, T.; Jenjaroenpun, P.; Chopjitt, P.; Boueroy, P.; Akeda, Y.; Hamada, S. Genomic Analysis of Aeromonas veronii C198, a Novel Mcr-3.41-Harboring Isolate from a Patient with Septicemia in Thailand. Pathogens 2020, 9, 1031. https://doi.org/10.3390/pathogens9121031
Hatrongjit R, Kerdsin A, Takeuchi D, Wongsurawat T, Jenjaroenpun P, Chopjitt P, Boueroy P, Akeda Y, Hamada S. Genomic Analysis of Aeromonas veronii C198, a Novel Mcr-3.41-Harboring Isolate from a Patient with Septicemia in Thailand. Pathogens. 2020; 9(12):1031. https://doi.org/10.3390/pathogens9121031
Chicago/Turabian StyleHatrongjit, Rujirat, Anusak Kerdsin, Dan Takeuchi, Thidathip Wongsurawat, Piroon Jenjaroenpun, Peechanika Chopjitt, Parichart Boueroy, Yukihiro Akeda, and Shigeyuki Hamada. 2020. "Genomic Analysis of Aeromonas veronii C198, a Novel Mcr-3.41-Harboring Isolate from a Patient with Septicemia in Thailand" Pathogens 9, no. 12: 1031. https://doi.org/10.3390/pathogens9121031
APA StyleHatrongjit, R., Kerdsin, A., Takeuchi, D., Wongsurawat, T., Jenjaroenpun, P., Chopjitt, P., Boueroy, P., Akeda, Y., & Hamada, S. (2020). Genomic Analysis of Aeromonas veronii C198, a Novel Mcr-3.41-Harboring Isolate from a Patient with Septicemia in Thailand. Pathogens, 9(12), 1031. https://doi.org/10.3390/pathogens9121031






