Genetic Analysis of Zoonotic Gastrointestinal Protozoa and Microsporidia in Shelter Cats in South Korea
Abstract
1. Introduction
2. Results
2.1. nPCR and Molecular Identification
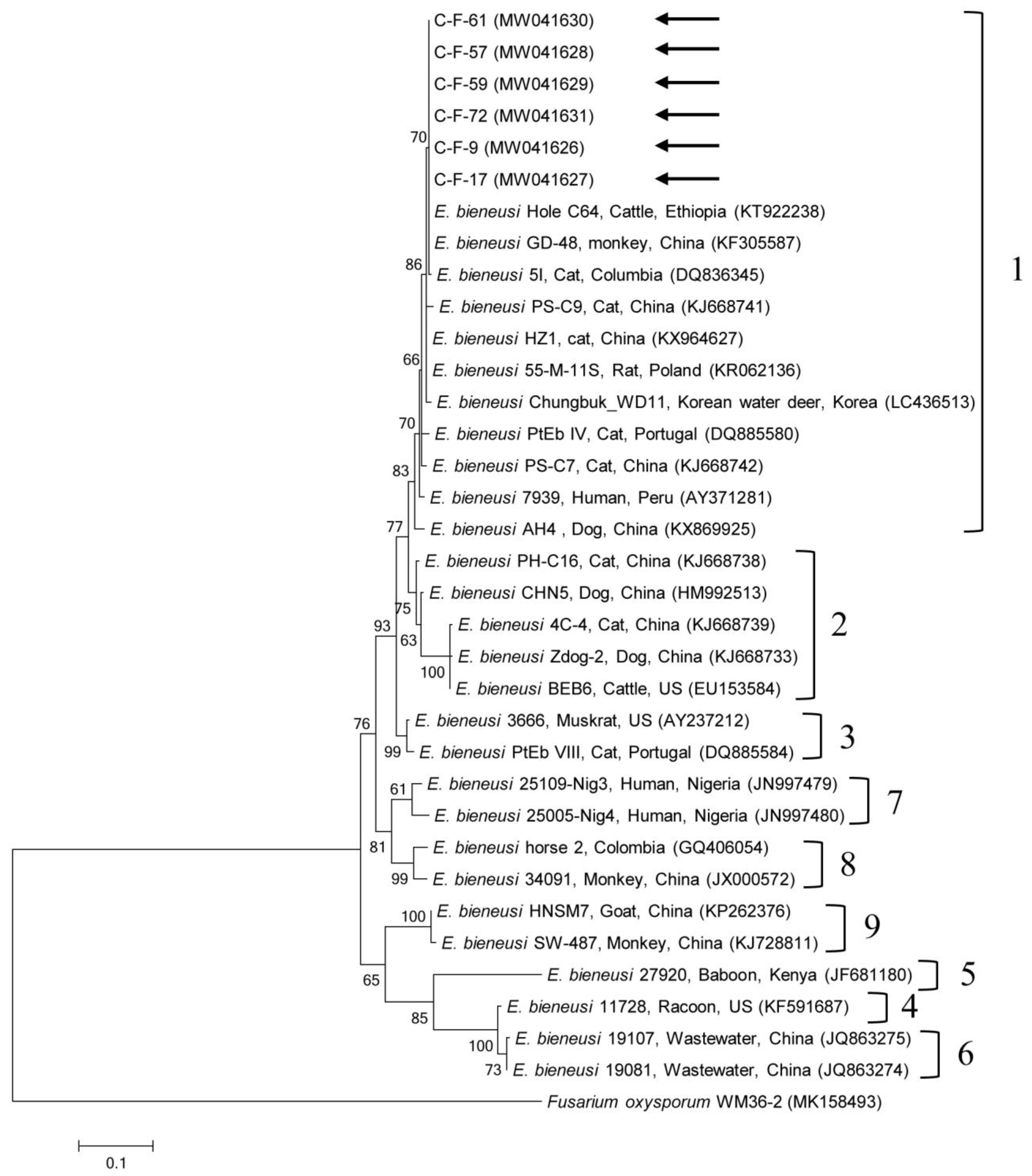
2.2. Molecular and Phylogenetic Analyses
3. Discussion
4. Materials and Methods
4.1. Sample Size and Collection
4.2. DNA Extraction and PCR
4.3. DNA Cloning
4.4. DNA Sequencing and Phylogenetic Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gerhold, R.W.; Jessup, D.A. Zoonotic diseases associated with free-roaming cats. Zoonoses. Public Health 2013, 60, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jin, Y.; Wu, W.; Li, P.; Wang, L.; Li, N.; Feng, Y.; Xiao, L. Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai, China. Parasit. Vectors 2016, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.; Munoz, M.; Gómez, N.; Tabares, J.; Segura, L.; Salazar, Á.; Restrepo, C.; Ruíz, M.; Reyes, P.; Qian, Y.; et al. Molecular Epidemiology of Giardia, Blastocystis and Cryptosporidium among indigenous children from the Colombian amazon basin. Front. Microbiol. 2017, 8, 248. [Google Scholar] [CrossRef]
- Reh, L.; Muadica, A.S.; Köster, P.C.; Balasegaram, S.; Verlander, N.Q.; Chércoles, E.R.; Carmena, D. Substantial prevalence of enteroparasites Cryptosporidium spp., Giardia duodenalis and Blastocystis sp. in asymptomatic schoolchildren in Madrid, Spain, November 2017 to June 2018. Euro. Surveill. 2019, 24, 1900241. [Google Scholar] [CrossRef]
- Wallace, J.L.; Levy, J.K. Population characteristics of feral cats admitted to seven trap-neuter-return programs in the United States. J. Feline Med. Surg. 2006, 8, 279–284. [Google Scholar] [CrossRef]
- Lee, S.E.; Kim, J.Y.; Kim, T.A.; Cho, S.H.; Ahn, H.J.; Woo, H.M.; Lee, W.J.; Nam, H.W. Prevalence of Toxoplasma gondii infection in stray and household cats in regions of Seoul, Korea. Korean J. Parasitol. 2010, 48, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Taetzsch, S.J.; Gruszynski, K.R.; Bertke, A.S.; Dubey, J.P.; Monti, K.A.; Zajac, A.M.; Lindsay, D.S. Prevalence of zoonotic parasites in feral cats of Central Virginia, USA. Zoonoses Public Health 2018, 65, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulou, D.; Claerebout, E.; Arvanitis, D.; Ligda, P.; Voutzourakis, N.; Casaert, S.; Sotiraki, S. Abundance, zoonotic potential and risk factors of intestinal parasitism amongst dog and cat populations: The scenario of Crete, Greece. Parasit. Vectors 2017, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Gates, M.C.; Nolan, T.J. Endoparasite prevalence and recurrence across different age groups of dogs and cats. Vet. Parasitol. 2009, 166, 153–158. [Google Scholar] [CrossRef]
- CDC. Outbreak of cryptosporidiosis associated with a splash park—Idaho, 2007. MMWR. Morb. Mortal. Wkly. Rep. 2009, 58, 615–618. [Google Scholar]
- Widmer, G.; Köster, P.C.; Carmena, D. Cryptosporidium hominis infections in non-human animal species: Revisiting the concept of host specificity. Int. J. Parasitol. 2020, 50, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Ying, J.L.; Monis, P.; Ryan, U. Molecular characterisation of Cryptosporidium and Giardia in cats (Felis catus) in Western Australia. Exp. Parasitol. 2015, 155, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; VanBik, D.; Kim, H.Y.; Lee, Y.R.; Kim, J.W.; Chae, M.; Oh, S.I.; Goo, Y.K.; Kwon, O.D.; Kwak, D. Multilocus typing of Cryptosporidium spp. in young calves with diarrhea in Korea. Vet. Parasitol. 2016, 229, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, X.; Gu, Y.; Liu, J.; Luo, J. Prevalence of Cryptosporidium, Giardia, Blastocystis, and trichomonads in domestic cats in East China. J. Vet. Med. Sci. 2019, 81, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Beser, J.; Toresson, L.; Eitrem, R.; Troell, K.; Winiecka-Krusnell, J.; Lebbad, M. Possible zoonotic transmission of Cryptosporidium felis in a household. Infect. Ecol. Epidemiol. 2015, 5, 28463. [Google Scholar] [CrossRef]
- De Lucio, A.; Bailo, B.; Aguilera, M.; Cardona, G.A.; Fernández-Crespo, J.C.; Carmena, D. No molecular epidemiological evidence supporting household transmission of zoonotic Giardia duodenalis and Cryptosporidium spp. from pet dogs and cats in the province of Álava, Northern Spain. Acta Trop. 2017, 170, 48–56. [Google Scholar] [CrossRef]
- Heyworth, M.F. Giardia duodenalis genetic assemblages and hosts. Parasite 2016, 23, 13. [Google Scholar] [CrossRef]
- CDC. Pathogen & Environment, giardia, Parasites. Available online: https://www.cdc.gov/parasites/giardia/pathogen.html (accessed on 1 June 2020).
- Kim, H.Y.; Lee, H.; Lee, S.H.; Seo, M.G.; Yi, S.; Kim, J.W.; Kim, C.H.; Lee, Y.R.; So, B.; Kwon, O.D.; et al. Multilocus genotyping and risk factor analysis of Giardia duodenalis in dogs in Korea. Acta Trop. 2019, 199, 105113. [Google Scholar] [CrossRef]
- Lee, H.; Jung, B.; Lim, J.S.; Seo, M.G.; Lee, S.H.; Choi, K.H.; Hwang, M.H.; Kim, T.H.; Kwon, O.D.; Kwak, D. Multilocus genotyping of Giardia duodenalis from pigs in Korea. Parasitol. Int. 2020, 78, 102154. [Google Scholar] [CrossRef]
- Esmailikia, L.; Ebrahimzade, E.; Shayan, P.; Amininia, N. Detection of small number of Giardia in biological materials prepared from stray dogs. Acta Parasitol. 2017, 62, 733–738. [Google Scholar] [CrossRef]
- Rehbein, S.; Klotz, C.; Ignatius, R.; Müller, E.; Aebischer, A.; Kohn, B. Giardia duodenalis in small animals and their owners in Germany: A pilot study. Zoonoses Public Health 2019, 66, 117–124. [Google Scholar] [CrossRef]
- Tan, K.S. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Stensvold, C.R.; Rajilić-Stojanović, M.; Heilig, H.G.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Clark, C.G. Pre-empting pandora’s box: Blastocystis subtypes revisited. Trends. Parasitol. 2020, 36, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Cian, A.; El Safadi, D.; Osman, M.; Moriniere, R.; Gantois, N.; Benamrouz-Vanneste, S.; Delgado-Viscogliosi, P.; Guyot, K.; Li, L.L.; Monchy, S.; et al. Molecular epidemiology of Blastocystis sp. in various animal groups from two french zoos and evaluation of potential zoonotic risk. PLoS ONE 2017, 12, e0169659. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, S.H.; Seo, M.G.; Kim, H.Y.; Kim, J.W.; Lee, Y.R.; Kim, J.H.; Kwon, O.D.; Kwak, D. Occurrence and genetic diversity of Blastocystis in Korean cattle. Vet. Parasitol. 2018, 258, 70–73. [Google Scholar] [CrossRef]
- Lee, H.; Seo, M.G.; Oem, J.K.; Kim, Y.S.; Lee, S.Y.; Kim, J.; Jeong, H.; Jheong, W.H.; Kim, Y.; Lee, W.J.; et al. Molecular detection and subtyping of Blastocystis detected in wild boars (Sus scrofa) in South Korea. J. Wildl. Dis. 2020, 56, 662–666. [Google Scholar] [CrossRef]
- Kim, M.J.; Won, E.J.; Kim, S.H.; Shin, J.H.; Chai, J.Y. Molecular detection and subtyping of human Blastocystis and the clinical implications: Comparisons between diarrheal and non-diarrheal groups in Korean populations. Korean J. Parasitol. 2020, 58, 321–326. [Google Scholar] [CrossRef]
- Nagel, R.; Cuttell, L.; Stensvold, C.R.; Mills, P.C.; Bielefeldt-Ohmann, H.; Traub, R.J. Blastocystis subtypes in symptomatic and asymptomatic family members and pets and response to therapy. Intern. Med. J. 2012, 42, 1187–1195. [Google Scholar] [CrossRef]
- Ruaux, C.G.; Stang, B.V. Prevalence of blastocystis in shelter-resident and client-owned companion animals in the US Pacific Northwest. PLoS ONE 2014, 9, e107496. [Google Scholar] [CrossRef]
- Katsumata, M.; Yoshikawa, H.; Tokoro, M.; Mizuno, T.; Nagamoto, T.; Hendarto, J.; Asih, P.B.S.; Rozi, I.E.; Kimata, I.; Takami, K.; et al. Molecular phylogeny of Blastocystis isolates from wild rodents captured in Indonesia and Japan. Parasitol. Res. 2018, 117, 2841–2846. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Alfellani, M.A.; Nørskov-Lauritsen, S.; Prip, K.; Victory, E.L.; Maddox, C.; Nielsen, H.V.; Clark, C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009, 39, 473–479. [Google Scholar] [CrossRef]
- Wang, W.; Owen, H.; Traub, R.J.; Cuttell, L.; Inpankaew, T.; Bielefeldt-Ohmann, H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet. Parasitol. 2014, 203, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Paulos, S.; Köster, P.C.; de Lucio, A.; Hernández-de-Mingo, M.; Cardona, G.A.; Fernández-Crespo, J.C.; Stensvold, C.R.; Carmena, D. Occurrence and subtype distribution of Blastocystis sp. in humans, dogs and cats sharing household in northern Spain and assessment of zoonotic transmission risk. Zoonoses Public Health 2018, 65, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Santín, M.; Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011, 90, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.; Kim, S.; Han, J.I.; Na, K.J. Prevalence and genotypes of Enterocytozoon bieneusi in wildlife in Korea: A public health concern. Parasit. Vectors 2019, 12, 160. [Google Scholar] [CrossRef]
- Lee, S.H.; Oem, J.K.; Lee, S.M.; Son, K.; Jo, S.D.; Kwak, D. Molecular detection of Enterocytozoon bieneusi from bats in South Korea. Med. Mycol. 2018, 56, 1033–1037. [Google Scholar] [CrossRef]
- Dashti, A.; Santín, M.; Cano, L.; de Lucio, A.; Bailo, B.; de Mingo, M.H.; Köster, P.C.; Fernández-Basterra, J.A.; Aramburu-Aguirre, J.; López-Molina, N.; et al. Occurrence and genetic diversity of Enterocytozoon bieneusi (Microsporidia) in owned and sheltered dogs and cats in Northern Spain. Parasitol. Res. 2019, 118, 2979–2987. [Google Scholar] [CrossRef]
- Li, X.; Xiao, L. Multilocus Sequence Typing and Population Genetic Analysis of Enterocytozoon bieneusi: Host Specificity and Its Impacts on Public Health. Front. Genet. 2019, 10, 307. [Google Scholar] [CrossRef]
- Frenkel, J.K.; Dubey, J.P. Toxoplasmosis and its prevention in cats and man. J. Infect. Dis. 1972, 126, 664–673. [Google Scholar] [CrossRef]
- Howe, D.K.; Sibley, L.D. Toxoplasma gondii comprises three clonal lineages: Correlation of parasite genotype with human disease. J. Infect. Dis. 1995, 172, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.S.; Ahn, A.J.; Park, S.I.; Sohn, W.M.; Shim, J.H.; Shin, S.S. Excretion of Toxoplasma gondii oocysts from Feral Cats in Korea. Korean J. Parasitol. 2019, 57, 665–670. [Google Scholar] [CrossRef]
- Jung, B.K.; Lee, S.E.; Lim, H.; Cho, J.; Kim, D.G.; Song, H.; Kim, M.J.; Shin, E.H.; Chai, J.Y. Toxoplasma gondii B1 gene detection in feces of stray cats around Seoul, Korea and genotype analysis of two laboratory-passaged isolates. Korean J. Parasitol. 2015, 53, 259–263. [Google Scholar] [CrossRef]
- Park, Y.; Noh, J.; Seo, H.J.; Kim, K.H.; Min, S.; Yoo, M.S.; Yun, B.R.; Kim, J.H.; Choi, E.J.; Cheon, D.S.; et al. Seroprevalence and B1 gene phylogeny of Toxoplasma gondii of dogs and cats in republic of Korea. Korean J. Parasitol. 2020, 58, 257–265. [Google Scholar] [CrossRef]
- Jung, B.K.; Song, H.; Lee, S.E.; Kim, M.J.; Cho, J.; Shin, E.H.; Chai, J.Y. Seroprevalence and risk factors of Toxoplasma gondii Infection among cat sitters in Korea. Korean J. Parasitol. 2017, 55, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Thrusfield, M. Veterinary Epidemiology, 3rd ed.; Blackwell Publishing: Oxford, UK, 2005. [Google Scholar]
- Cheun, H.I.; Choi, T.K.; Chung, G.T.; Cho, S.H.; Lee, Y.H.; Kimata, I.; Kim, T.S. Genotypic characterization of Cryptosporidium oocysts isolated from healthy people in three different counties of Korea. J. Vet. Med. Sci. 2007, 69, 1099–1101. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Beck, R.; Lalle, M.; Marinculic, A.; Pozio, E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008, 38, 1523–1531. [Google Scholar] [CrossRef]
- Cacciò, S.M.; De Giacomo, M.; Pozio, E. Sequence analysis of the beta-giardin gene and development of a PCR-RFLP assay to genotype Giardia duodenalis cysts from human fecal samples. Int. J. Parasitol. 2002, 32, 1023–1030. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Sánchez, L.V.; Bautista, D.C.; Corredor, A.F.; Flórez, A.C.; Stensvold, C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014, 22, 223–228. [Google Scholar]
- Sulaiman, I.M.; Bern, C.; Gilman, R.; Cama, V.; Kawai, V.; Vargas, D.; Ticona, E.; Vivar, A.; Xiao, L. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. J. Eukaryot. Microbiol. 2003, 50, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Grigg, M.E.; Boothroyd, J.C. Rapid identification of virulent type 1 Strains of the protozoa pathogen Toxoplasma gondii by PCR-restriction fragment length polymorphism analysis at the B1 gene. J. Clin. Microbiol. 2001, 39, 398–400. [Google Scholar] [CrossRef] [PubMed]

| Category | No. Tested | Cryptosporidium felis | Giardia duodenalis | Blastocystis sp. | Enterocytozoon bieneusi | Toxoplasma gondii | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive (%) | 95% CI | Positive (%) | 95% CI | Positive (%) | 95% CI | Positive (%) | 95% CI | Positive (%) | 95% CI | ||||
| Sex | Female | 96 | 1 (1.0) | 0–3.1 | 3 (3.1) | 0–6.6 | 1 (1.0) | 0–3.1 | 4 (4.2) | 0.2–8.2 | 1 (1.0) | 0–3.1 | 10 (10.4) |
| Male | 62 | 0 | 0 | 3 (4.8) | 0–10.2 | 0 | 0 | 2 (3.2) | 0–7.6 | 1 (1.6) | 0–4.8 | 6 (9.7) | |
| Age (months) | ≤6 | 50 | 1 (2.0) | 0–5.9 | 3 (6.0) | 0–12.6 | 1 (2.0) | 0–5.9 | 2 (4.0) | 0–9.4 | 1 (2.0) | 0–5.9 | 8 (16.0) |
| >6 | 108 | 0 | 0 | 3 (2.8) | 0–5.9 | 0 | 0 | 4 (3.7) | 0.1–7.3 | 1 (0.9) | 0–2.7 | 8 (7.4) | |
| Total | 158 | 1 (0.6) | 0–1.9 | 6 (3.8) | 0.8–6.8 | 1 (0.6) | 0–1.9 | 2 (1.3) | 0.8–6.8 | 2 (1.3) | 0–3.0 | 16 (10.1) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwak, D.; Seo, M.-G. Genetic Analysis of Zoonotic Gastrointestinal Protozoa and Microsporidia in Shelter Cats in South Korea. Pathogens 2020, 9, 894. https://doi.org/10.3390/pathogens9110894
Kwak D, Seo M-G. Genetic Analysis of Zoonotic Gastrointestinal Protozoa and Microsporidia in Shelter Cats in South Korea. Pathogens. 2020; 9(11):894. https://doi.org/10.3390/pathogens9110894
Chicago/Turabian StyleKwak, Dongmi, and Min-Goo Seo. 2020. "Genetic Analysis of Zoonotic Gastrointestinal Protozoa and Microsporidia in Shelter Cats in South Korea" Pathogens 9, no. 11: 894. https://doi.org/10.3390/pathogens9110894
APA StyleKwak, D., & Seo, M.-G. (2020). Genetic Analysis of Zoonotic Gastrointestinal Protozoa and Microsporidia in Shelter Cats in South Korea. Pathogens, 9(11), 894. https://doi.org/10.3390/pathogens9110894





