The Use of Hyperimmune Chicken Reference Sera Is Not Appropriate for the Validation of Influenza Pseudotype Neutralization Assays
Abstract
1. Introduction
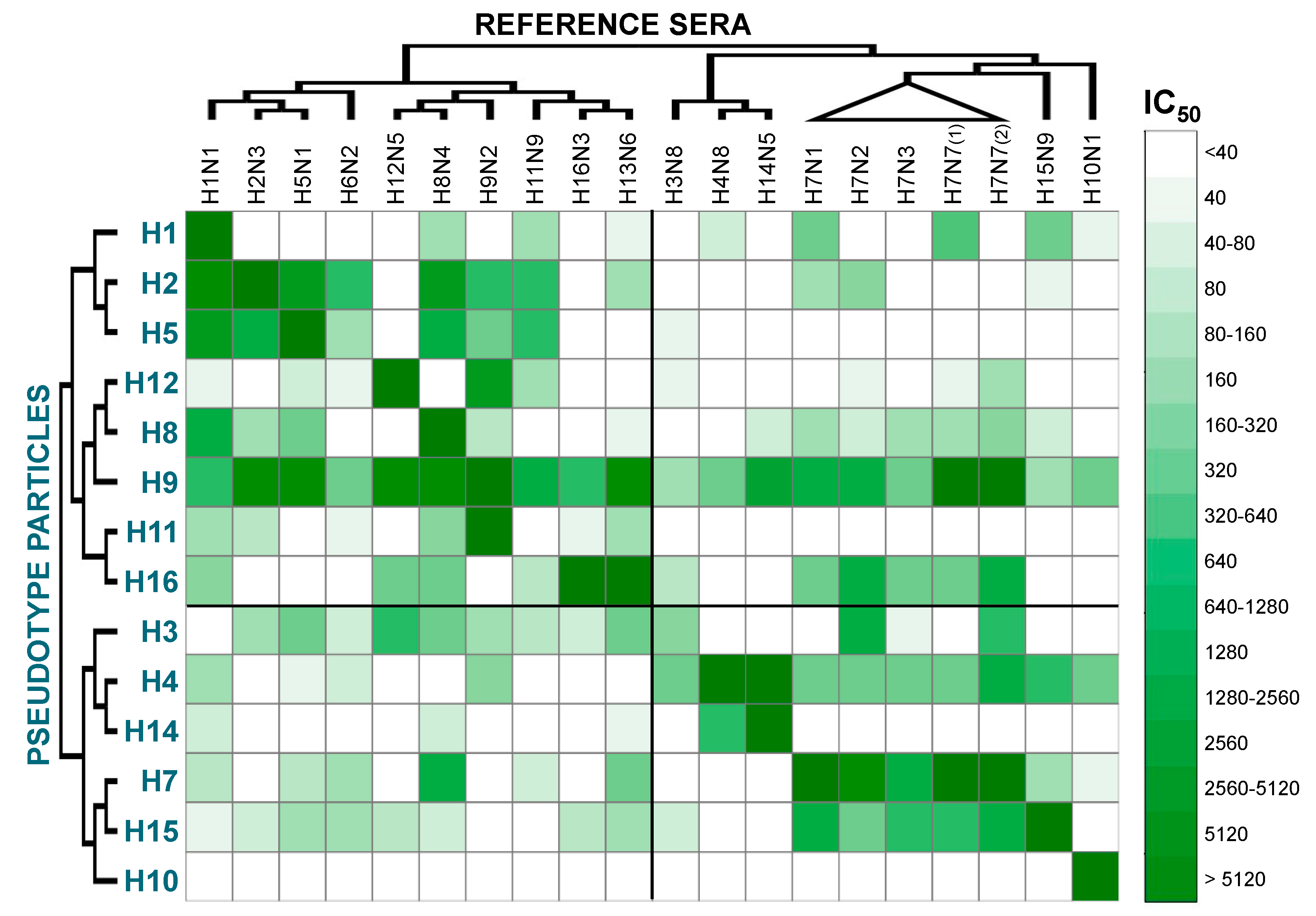
2. Results and Discussion
3. Materials and Methods
3.1. Reference Sera
3.2. Pseudotype Particles and Pseudotype Particle Neutralization Assays
3.3. Bioinformatic Analysis
3.4. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Note
- Trombetta, C.; Perini, D.; Mather, S.; Temperton, N.; Montomoli, E. Overview of Serological Techniques for Influenza Vaccine Evaluation: Past, Present and Future. Vaccines 2014, 2, 707–734. [Google Scholar] [CrossRef] [PubMed]
- Carnell, G.W.; Ferrara, F.; Grehan, K.; Thompson, C.P.; Temperton, N.J. Pseudotype-based neutralization assays for influenza: A systematic analysis. Front. Immunol. 2015, 6, 161. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, F.; Molesti, E.; Temperton, N. The application of pseudotypes to influenza pandemic preparedness. Future Virol. 2015, 10, 731–749. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.-M.; Lagarde, N.; Ma, E.S.K.; de Jong, M.D.; Peiris, J.S.M. Optimization and evaluation of an influenza A (H5) pseudotyped lentiviral particle-based serological assay. J. Clin. Virol. 2010, 47, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xie, H.; Ye, Z.; Vassell, R.; Weiss, C.D. Characterization of lentiviral pseudotypes with influenza H5N1 hemagglutinin and their performance in neutralization assays. J. Virol. Methods 2010, 165, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Alberini, I.; Del Tordello, E.; Fasolo, A.; Temperton, N.J.; Galli, G.; Gentile, C.; Montomoli, E.; Hilbert, A.K.; Banzhoff, A.; Del Giudice, G.; et al. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine 2009, 27, 5998–6003. [Google Scholar] [CrossRef] [PubMed]
- Temperton, N.J.; Hoschler, K.; Major, D.; Nicolson, C.; Manvell, R.; Hien, V.M.; Ha do, Q.; de Jong, M.; Zambon, M.; Takeuchi, Y.; et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Respir. Viruses 2007, 1, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.H. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. 1998, 17, 469–526. [Google Scholar] [CrossRef] [PubMed]
- The United States Pharmacopeial Convention. <1033> Biological Assay Validation. 2010.
- Gray, J.J.; Wreghitt, T.G.; McKee, T.A.; McIntyre, P.; Roth, C.E.; Smith, D.J.; Sutehall, G.; Higgins, G.; Geraghty, R.; Whetstone, R. Internal quality assurance in a clinical virology laboratory. II. Internal quality control. J. Clin. Pathol. 1995, 48, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.M.; Gaines-Das, R.E.; Taylor, J.; Chakraverty, P. Comparison of influenza serological techniques by international collaborative study. Vaccine 1994, 12, 167–174. [Google Scholar] [CrossRef]
- World Organization for Animal Health. Avian Influenza; OIE: Paris, France, 2012. [Google Scholar]
- Dwyer, D.E.; Smith, D.W.; Catton, M.G.; Barr, I.G. Laboratory diagnosis of human seasonal and pandemic influenza virus infection. Med. J. Aust. 2006, 185, S48–S53. [Google Scholar] [PubMed]
- Shahsavandi, S.; Salmanian, A.-H.; Ghorashi, S.A.; Masoudi, S.; Fotouhi, F.; Ebrahimi, M.M. Specific subtyping of influenza A virus using a recombinant hemagglutinin protein expressed in baculovirus. Mol. Biol. Rep. 2011, 38, 3293–3298. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Senne, D.A.; Suarez, D.L. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clin. Vaccine Immunol. 2006, 13, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.M.; Smith, D.J. Use of antigenic cartography in vaccine seed strain selection. Avian Dis. 2010, 54, 220–223. [Google Scholar] [CrossRef] [PubMed]
- De Jong, J.C.; Smith, D.J.; Lapedes, A.S.; Donatelli, I.; Campitelli, L.; Barigazzi, G.; van Reeth, K.; Jones, T.C.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; et al. Antigenic and Genetic Evolution of Swine Influenza A (H3N2) Viruses in Europe. J. Virol. 2007, 81, 4315–4322. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, F.; Molesti, E.; Böttcher-Friebertshäuser, E.; Cattoli, G.; Corti, D.; Scott, S.D.; Temperton, N.J. The human Transmembrane Protease Serine 2 is necessary for the production of Group 2 influenza A virus pseudotypes. J. Mol. Genet. Med. 2012, 7, 309–314. [Google Scholar] [CrossRef] [PubMed]
- GISAID: Global initiative on sharing all influenza data—From vision to reality. Euro Surveill. 2017, 22, 30494. [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]

| Antigen Strain Name | Subtype | HA Accession Number | HI Titre |
|---|---|---|---|
| A/duck/Italy/1447/2005 | H1N1 | HF563054.1 | 1:512 |
| A/duck/Germany/1215/1973 | H2N3 | CY014710.1 | 1:512 |
| A/psittacine/Italy/2873/2000 | H3N8 | GQ247846.1 * | 1:256 |
| A/cockatoo/England/1972 | H4N8 | GQ247847.1 * | 1:128 |
| A/turkey/Canada/1965 | H6N2 | GQ247851.1 * | 1:256 |
| A/turkey/Ontario/6118/1968 | H8N4 | CY014659.1 | 1:512 |
| A/mallard/Italy/3817-34/2005 | H9N2 | Not Applicable | 1:256 |
| A/ostrich/South Africa/2001 | H10N1 | GQ247860.1 * | 1:512 |
| A/duck/Memphis/546/1974 | H11N9 | AB292779.1 | 1:1024 |
| A/duck/Alberta/60/1976 | H12N5 | CY130078.1 | 1:128 |
| A/gull/Maryland/704/1977 | H13N6 | D90308.1 | 1:1024 |
| A/mallard/Gurjev/263/1982 | H14N5 | M35997 | 1:512 |
| A/shearwater/Australia/2576/1979 ** | H15N9 | CY130102.1 | 1:2048 |
| A/gull/Denmark/68110/2002 | H16N3 | GQ247872.1 * | 1:256 |
| Antigen Strain Name | Subtype | HA Accession Number | HI Titre |
|---|---|---|---|
| A/chicken/Scotland/1959 | H5N1 | CY015081.1 | Not available |
| A/African starling/England/983/1979 | H7N1 | AF202232.1 | Not available |
| A/chicken/Wales/1306/2007 | H7N2 | EF675618.1 | Not available |
| A/chicken/England/4054/2006 | H7N3 | EF467826.1 | Not available |
| A/England/268/1996 | H7N7 | AF028020.1 | Not available |
| A/turkey/England/647/1977 | H7N7 | AF202247.1 | Not available |
| Plasmid Backbone | HA | HA Accession Number |
|---|---|---|
| pI.18 | A/duck/Italy/1447/2005 H1 | HF563054.1 |
| pI.18 | A/Korea/426/1968 H2 | CY125846.1 |
| phCMV1 | A/duck/Germany/1215/1973 H2 | CY014710.1 |
| pI.18 | A/Udorn/307/1972 H3 | DQ508929.1 |
| phCMV1 | A/duck/Czechoslovakia/1956 H4 | D90302.1 |
| pI.18 | A/Vietnam/1194/2004 H5 | ABP51976.1 |
| pI.18 | A/chicken/Italy/1082/1999 H7 | ABR37396.1 |
| phCMV1 | A/turkey/Ontario/6118/1968 H8 | CY014659.1 |
| pI.18 | A/Hong Kong/1073/1999 H9 | AJ404626.1 |
| phCMV1 | A/chicken/Germany/N49 H10 | CY014671.1 |
| phCMV1 | A/duck/Memphis/546/1974 H11 | AB292779.1 |
| phCMV1 | A/duck/Alberta/60/1976 H12 | CY130078.1 |
| phCMV1 | A/gull/Maryland/704/1977 H13 | D90308.1 |
| phCMV1 | A/mallard/Astrakhan/263/1982 H14 | AB289335.1 |
| phCMV1 | A/shearwater/West Australia/2576/1979 H15 | CY130102.1 |
| phCMV1 | A/black-headed gull/Sweden/2/1999 H16 | AY684888.1 |
| Group | IC50 Values | Dilution Factor |
|---|---|---|
| 0 | <35 | <40 |
| 1 | 35–45 | 40 |
| 2 | 45–75 | 40–80 |
| 3 | 75–85 | 80 |
| 4 | 85–150 | 80–160 |
| 5 | 150–170 | 160 |
| 6 | 170–310 | 160–320 |
| 7 | 310–330 | 320 |
| 8 | 330–630 | 320–640 |
| 9 | 630–670 | 640 |
| 10 | 670–1270 | 640–1280 |
| 11 | 1270–1290 | 1280 |
| 12 | 1290–2550 | 1280–2560 |
| 13 | 2550–2570 | 2560 |
| 14 | 2570–5100 | 2560–5120 |
| 15 | 5100–5140 | 5120 |
| 16 | >5140 | >5120 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrara, F.; Molesti, E.; Scott, S.; Cattoli, G.; Temperton, N. The Use of Hyperimmune Chicken Reference Sera Is Not Appropriate for the Validation of Influenza Pseudotype Neutralization Assays. Pathogens 2017, 6, 45. https://doi.org/10.3390/pathogens6040045
Ferrara F, Molesti E, Scott S, Cattoli G, Temperton N. The Use of Hyperimmune Chicken Reference Sera Is Not Appropriate for the Validation of Influenza Pseudotype Neutralization Assays. Pathogens. 2017; 6(4):45. https://doi.org/10.3390/pathogens6040045
Chicago/Turabian StyleFerrara, Francesca, Eleonora Molesti, Simon Scott, Giovanni Cattoli, and Nigel Temperton. 2017. "The Use of Hyperimmune Chicken Reference Sera Is Not Appropriate for the Validation of Influenza Pseudotype Neutralization Assays" Pathogens 6, no. 4: 45. https://doi.org/10.3390/pathogens6040045
APA StyleFerrara, F., Molesti, E., Scott, S., Cattoli, G., & Temperton, N. (2017). The Use of Hyperimmune Chicken Reference Sera Is Not Appropriate for the Validation of Influenza Pseudotype Neutralization Assays. Pathogens, 6(4), 45. https://doi.org/10.3390/pathogens6040045







