Brighter Fluorescent Derivatives of UTI89 Utilizing a Monomeric vGFP
Abstract
:1. Introduction
2. Results and Discussion
2.1. New vsfGFP-9 Expressing Derivatives of UTI89 Are 10× Brighter Than Former GFP Expressing Strains
2.2. New Chromosomal vsfGFP-9 Construct Has No Fitness Defects during UTI Relative to Former GFP Expressing Strains
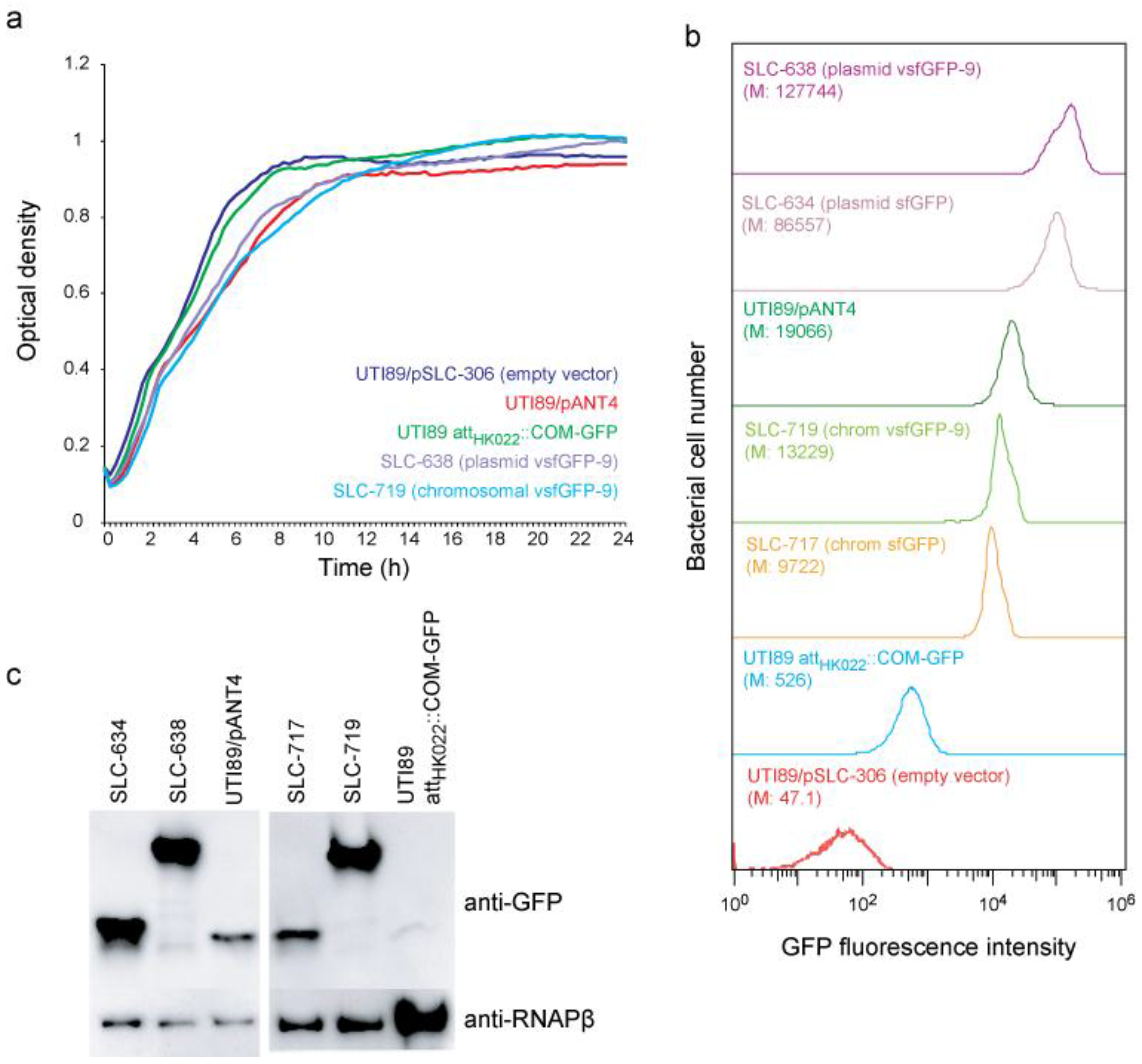
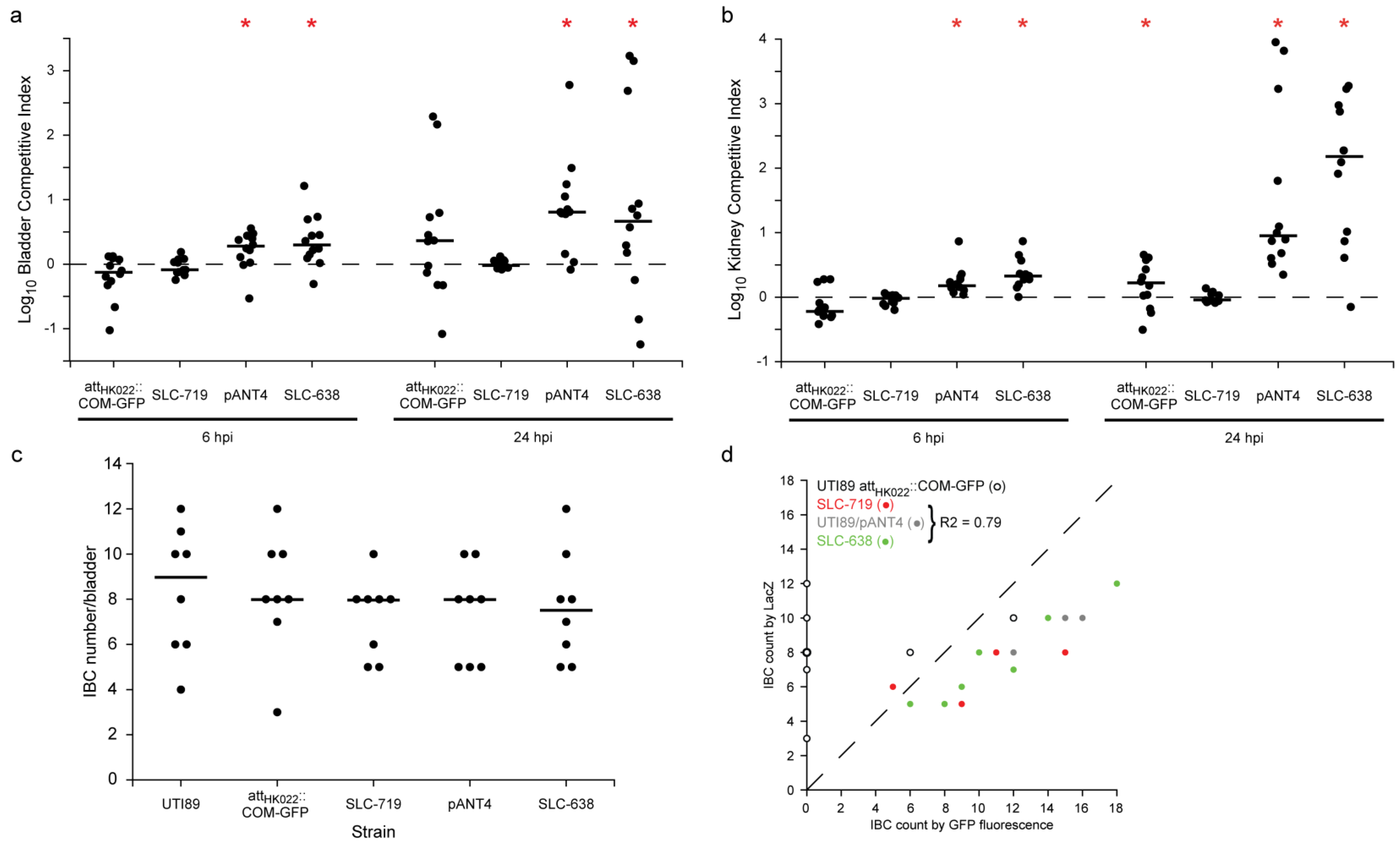
3. Experimental Section
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Valdivia, R.H.; Hromockyj, A.E.; Monack, D.; Ramakrishnan, L.; Falkow, S. Applications for green fluorescent protein (gfp) in the study of hostpathogen interactions. Gene 1996, 173, 47–52. [Google Scholar] [CrossRef]
- Hautefort, I.; Proença, M.J.; Hinton, J.C. Single-copy green fluorescent protein gene fusions allow accurate measurement of salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 2003, 69, 7480–7491. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Hautefort, I.; Hinton, J.C. Measurement of bacterial gene expression in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.K.; Hooton, T.M.; Martin, S.M.; Stamm, W.E.; Palermo, J.J.; Gordon, J.I.; Hultgren, S.J. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect. Immun. 2007, 75, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Hung, C.; Theriot, J.A.; Fletcher, D.A.; Anderson, G.G.; Footer, M.J.; Hultgren, S.J. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Mysorekar, I.U.; Hultgren, S.J. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. USA 2006, 103, 14170–14175. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Hung, C.-S.; Xu, J.; Reigstad, C.S.; Magrini, V.; Sabo, A.; Blasiar, D.; Bieri, T.; Meyer, R.R.; Ozersky, P. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. USA 2006, 103, 5977–5982. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Falkow, S. Constitutive and inducible green fluorescent protein expression in bartonella henselae. Infect. Immun. 1998, 66, 3964–3967. [Google Scholar] [PubMed]
- Wright, K.J.; Seed, P.C.; Hultgren, S.J. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 2005, 73, 7657–7668. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Lauer, S.R.; Hultgren, S.J.; Hunstad, D.A. Maturation of intracellular Escherichia coli communities requires sura. Infect. Immun. 2006, 74, 4793–4800. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Kirchhofer, A.; Helma, J.; Schmidthals, K.; Frauer, C.; Cui, S.; Karcher, A.; Pellis, M.; Muyldermans, S.; Casas-Delucchi, C.S.; Cardoso, M.C. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010, 17, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi, M.; Sun, G.; Gruter, A.; Lim, C.L.; Chee, Y.C.; Jung, G.; Jauch, R.; Wohland, T.; Chen, S.L. Rational structure-based design of bright gfp-based complexes with tunable dimerization. Angew. Chem. Int. Ed. Engl. 2015, 127, 14158–14162. [Google Scholar]
- Anderson, G.G.; Palermo, J.J.; Schilling, J.D.; Roth, R.; Heuser, J.; Hultgren, S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.S.; Dodson, K.W.; Hultgren, S.J. A murine model of urinary tract infection. Nat. Protoc. 2009, 4, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Orosz, A.; Boros, I.; Venetianer, P. Analysis of the complex transcription termination region of the Escherichia coli rrnb gene. Eur. J. Biochem. 1991, 201, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Dueber, J.E.; Leguia, M.; Wu, G.C.; Goler, J.A.; Arkin, A.P.; Keasling, J.D. Bglbricks: A flexible standard for biological part assembly. J. Biol. Eng. 2010, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Khetrapal, V.; Mehershahi, K.; Rafee, S.; Chen, S.; Lim, C.L.; Chen, S.L. A set of powerful negative selection systems for unmodified enterobacteriaceae. Nucleic Acids Res. 2015, 43, e83. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli k-12 using pcr products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eshaghi, M.; Mehershahi, K.S.; Chen, S.L. Brighter Fluorescent Derivatives of UTI89 Utilizing a Monomeric vGFP. Pathogens 2016, 5, 3. https://doi.org/10.3390/pathogens5010003
Eshaghi M, Mehershahi KS, Chen SL. Brighter Fluorescent Derivatives of UTI89 Utilizing a Monomeric vGFP. Pathogens. 2016; 5(1):3. https://doi.org/10.3390/pathogens5010003
Chicago/Turabian StyleEshaghi, Majid, Kurosh S. Mehershahi, and Swaine L. Chen. 2016. "Brighter Fluorescent Derivatives of UTI89 Utilizing a Monomeric vGFP" Pathogens 5, no. 1: 3. https://doi.org/10.3390/pathogens5010003
APA StyleEshaghi, M., Mehershahi, K. S., & Chen, S. L. (2016). Brighter Fluorescent Derivatives of UTI89 Utilizing a Monomeric vGFP. Pathogens, 5(1), 3. https://doi.org/10.3390/pathogens5010003





