E2 Tyrosine 102 Regulates MmuPV1 Pathogenesis In Vivo
Abstract
1. Introduction
2. Materials and Methods
3. Results
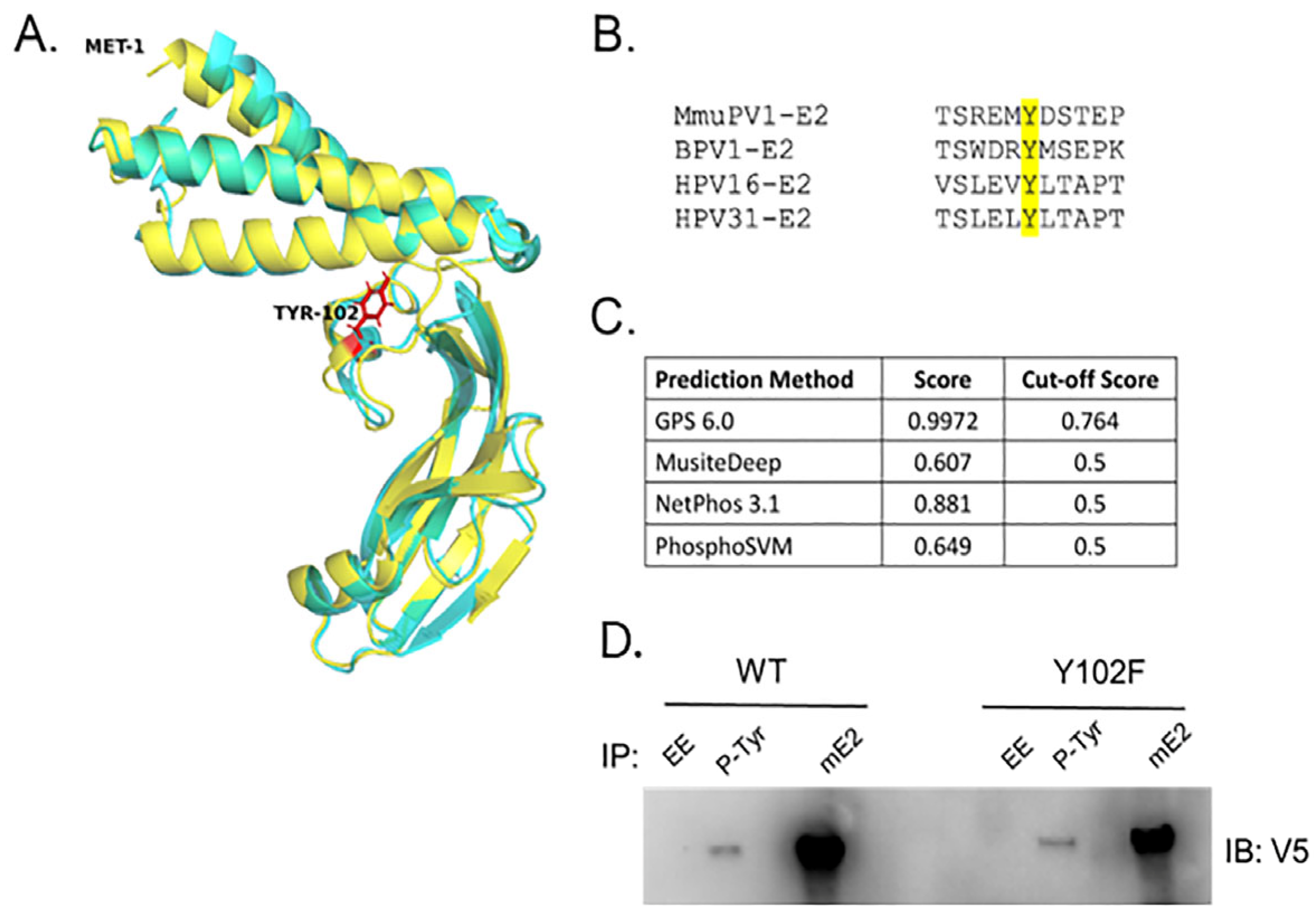
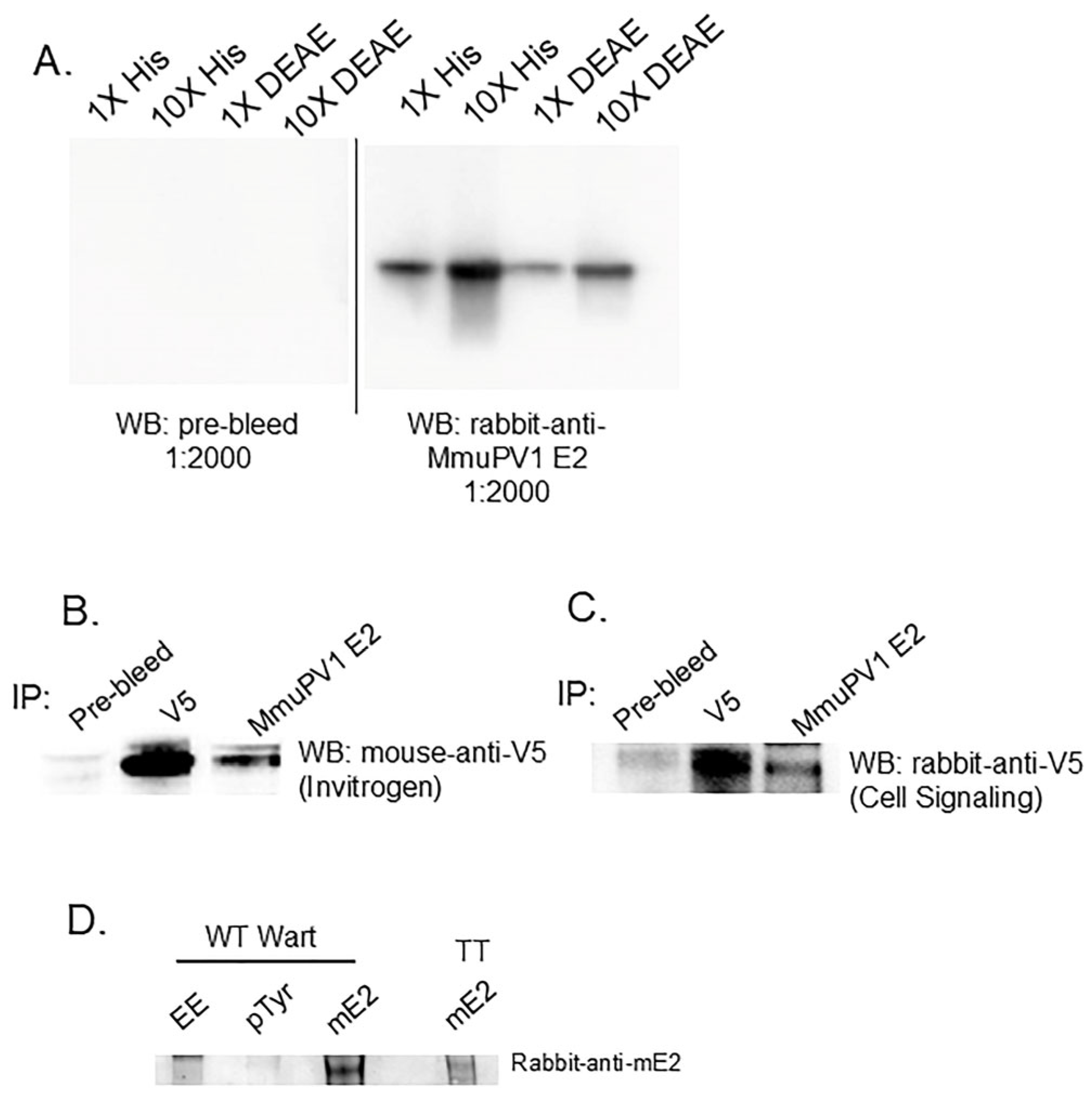
3.1. The MmuPV1 E2 Protein Is Phosphorylated at Tyrosine Residues
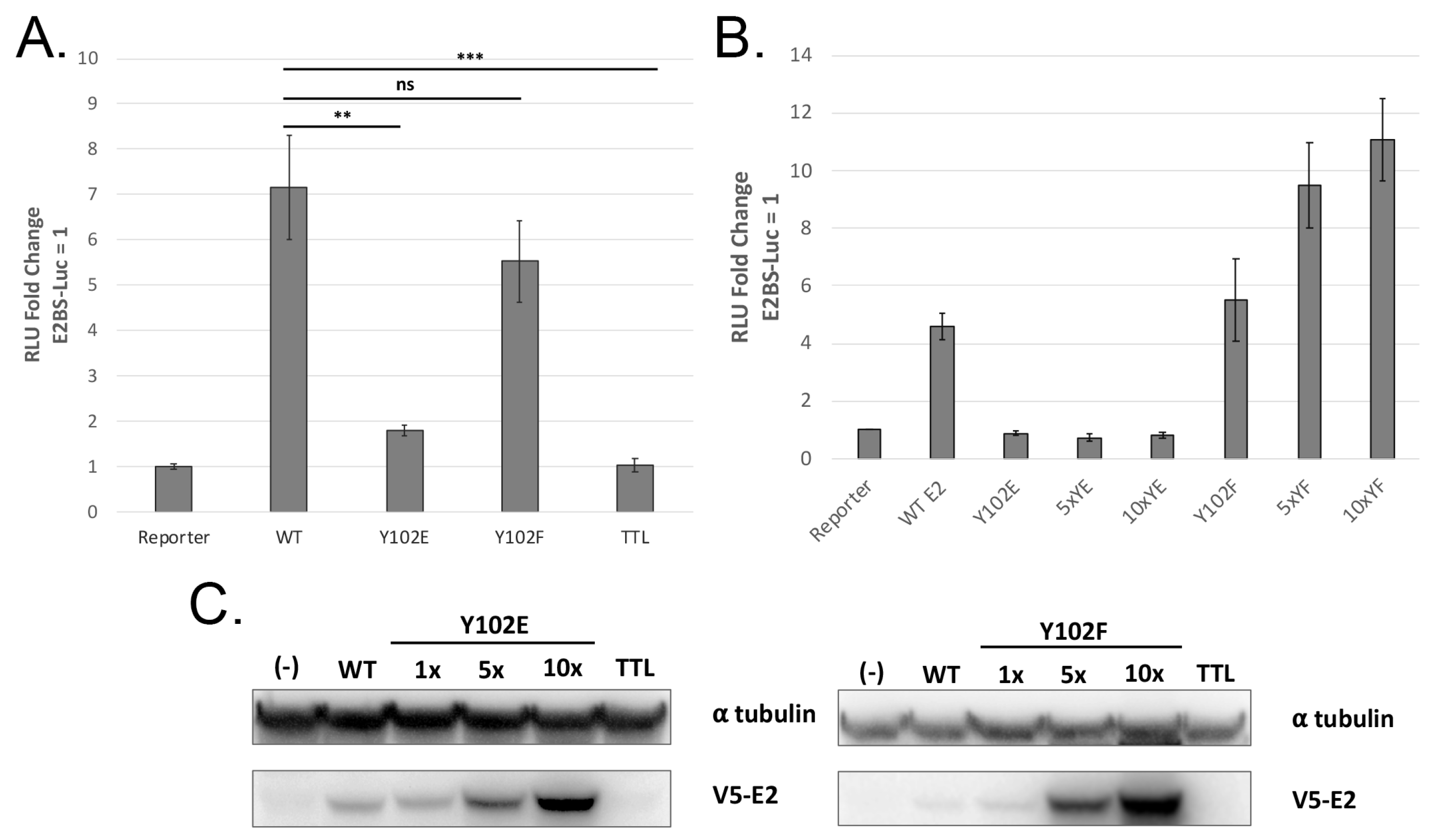
3.2. Y102E Is Transcription and Replication Defective
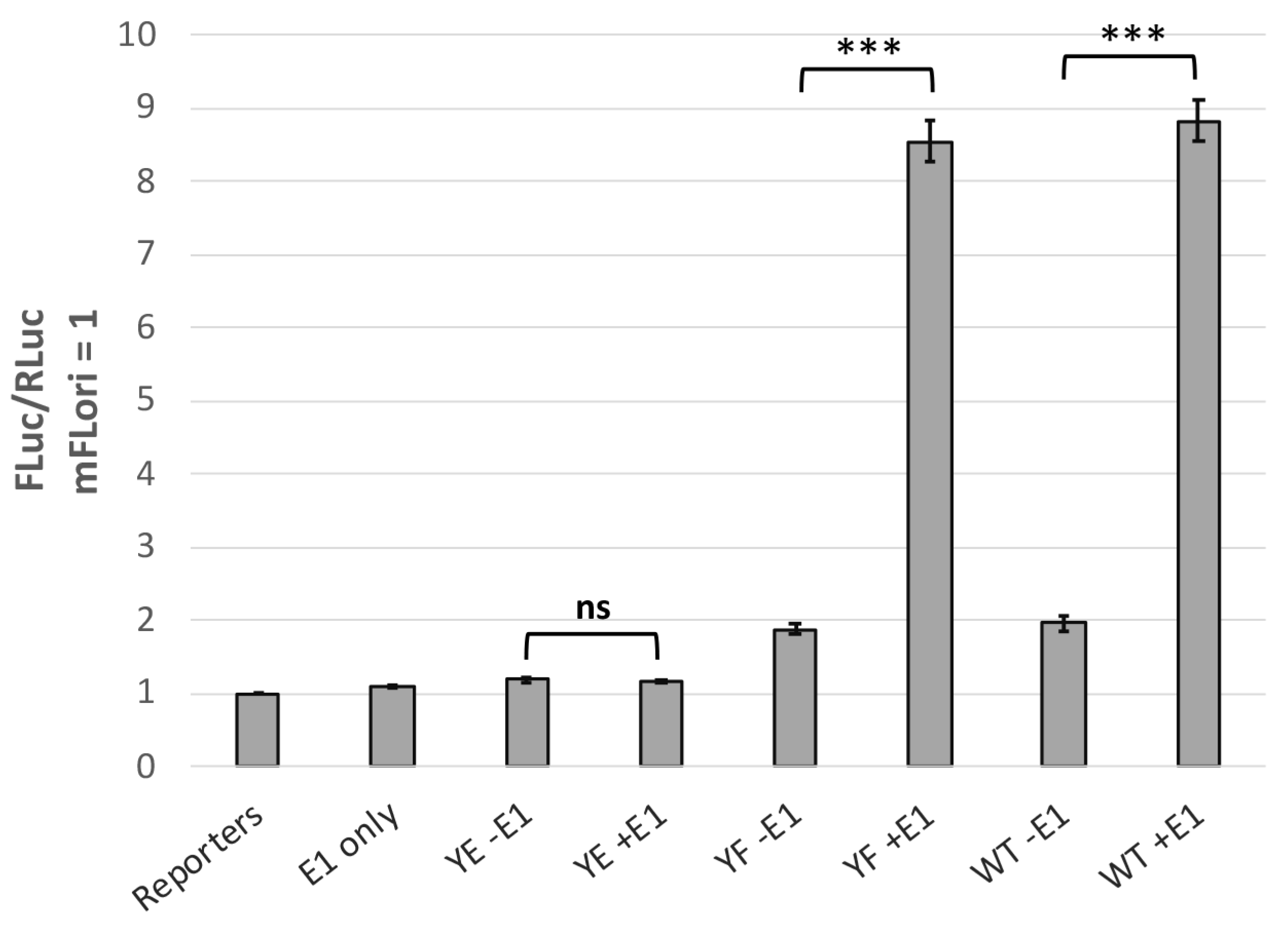
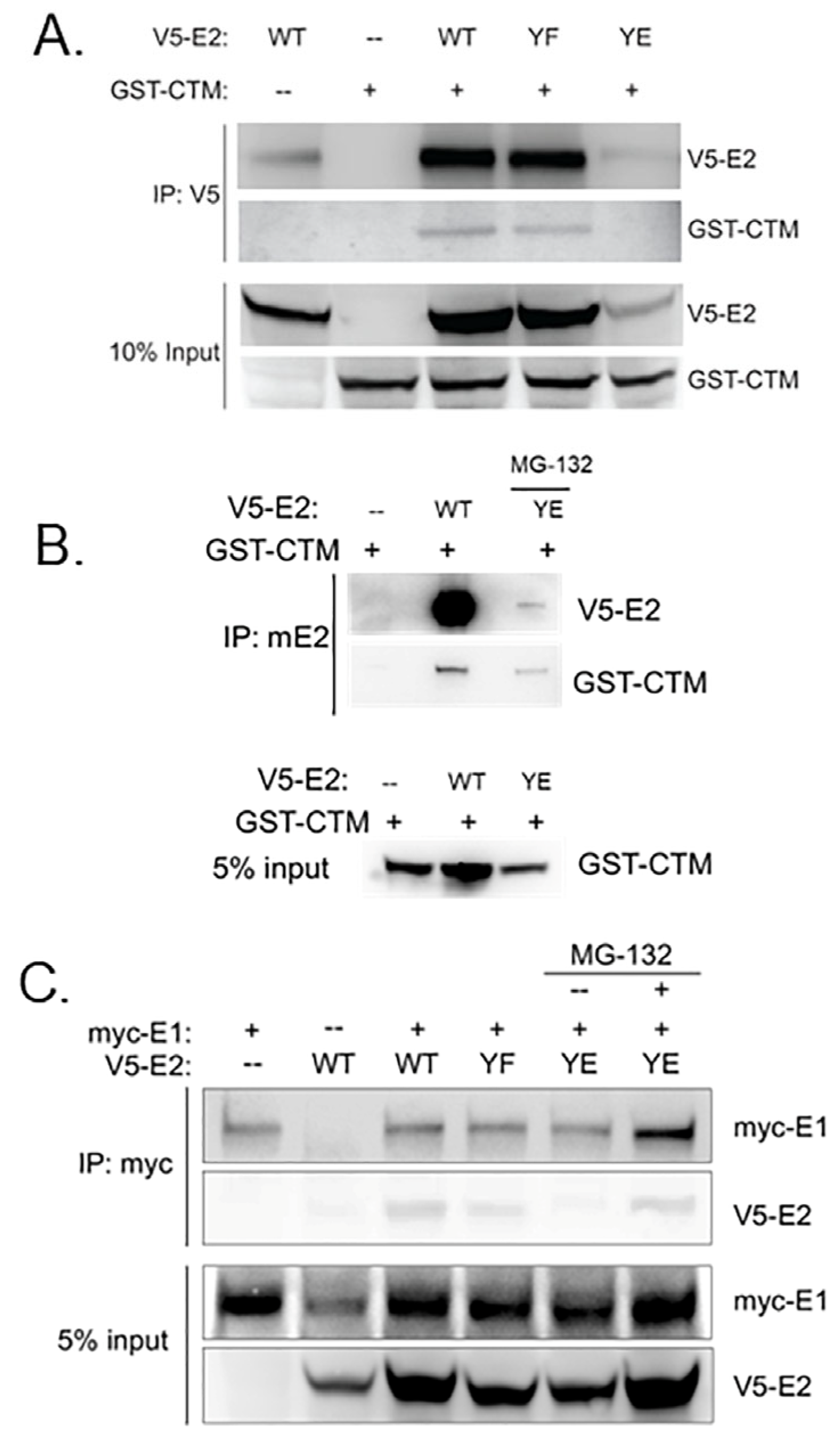
3.3. Y102 Affects E2 Protein–Protein Interactions
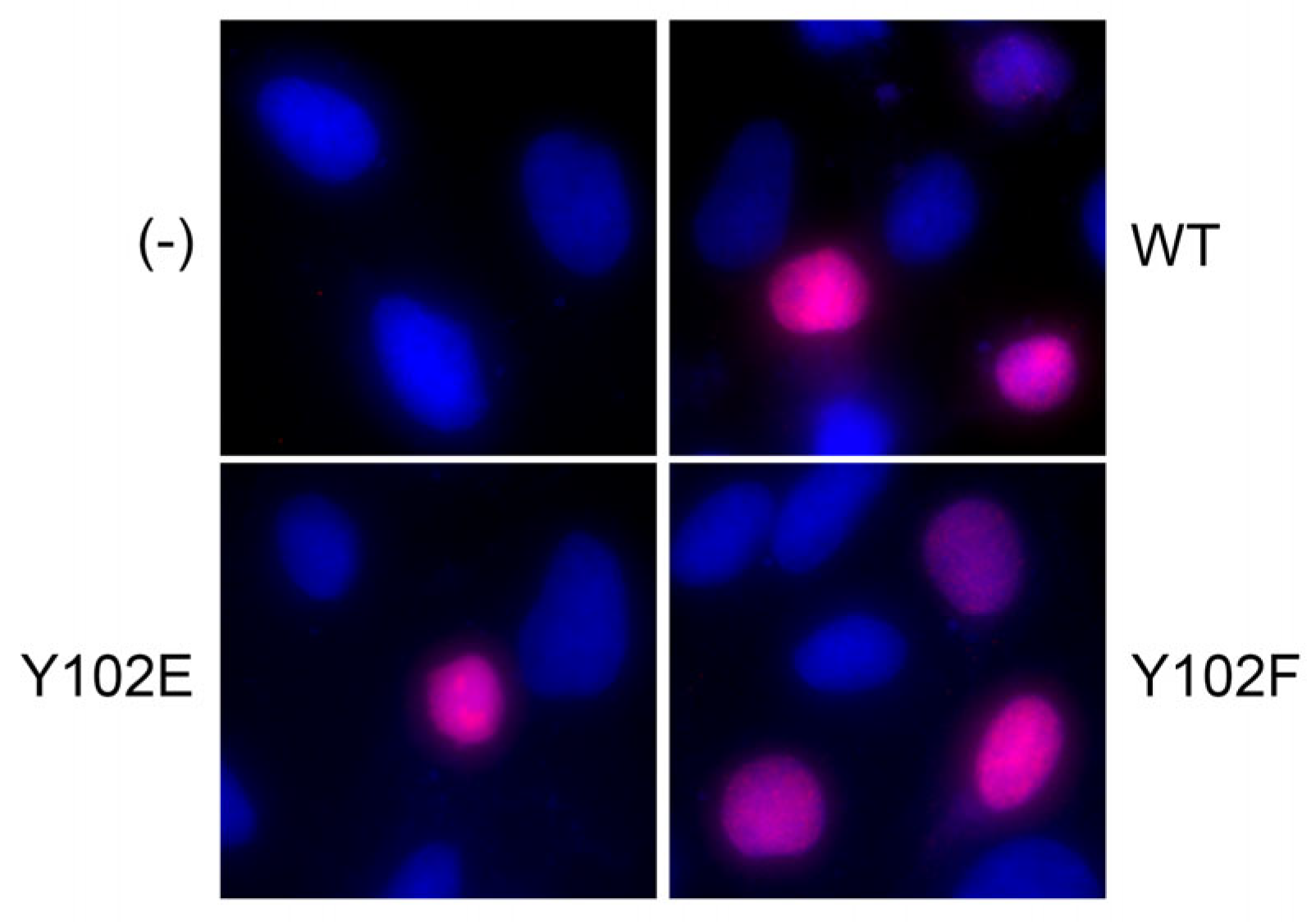
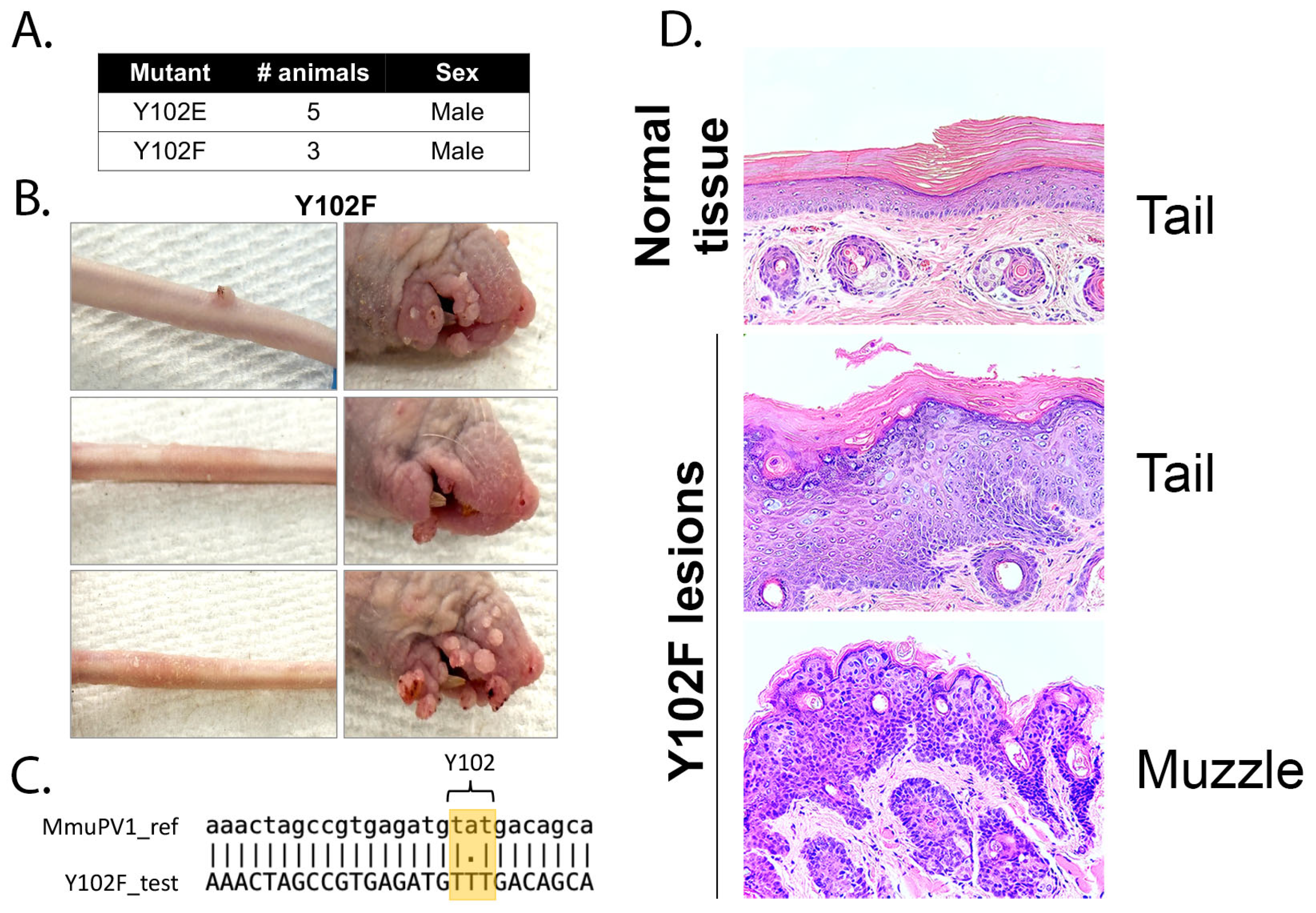
3.4. The E2 Y102F Mutant Genome Is Competent for Establishment and Production of Infectious Virus In Vivo
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stubenrauch, F.; Laimins, L.A. Human papillomavirus life cycle: Active and latent phases. Semin. Cancer Biol. 1999, 9, 379–386. [Google Scholar] [CrossRef]
- Roberts, J.N.; Buck, C.B.; Thompson, C.D.; Kines, R.; Bernardo, M.; Choyke, P.L.; Lowy, D.R.; Schiller, J.T. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 2007, 13, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Day, P.M.; Thompson, C.D.; Weisberg, A.S.; Schiller, J.T. The COPII Transport Complex Participates in HPV16 Infection. Viruses 2025, 17, 616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kazakov, T.; Popa, A.; DiMaio, D. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity. mBio 2014, 5, e01777-14. [Google Scholar] [CrossRef] [PubMed]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- Hoffmann, R.; Hirt, B.; Bechtold, V.; Beard, P.; Raj, K. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 2006, 80, 4431–4439. [Google Scholar] [CrossRef]
- Skiadopoulos, M.H.; McBride, A.A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 1998, 72, 2079–2088. [Google Scholar] [CrossRef]
- Lehman, C.W.; Botchan, M.R. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 1998, 95, 4338–4343. [Google Scholar] [CrossRef]
- Wu, S.Y.; Lee, A.Y.; Lai, H.T.; Zhang, H.; Chiang, C.M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell 2013, 49, 843–857. [Google Scholar] [CrossRef]
- Gonzalez, J.; Stoll, K.; DeSmet, M.; Androphy, E.J. A conserved cysteine in the DNA-binding domain of MmuPV1 E2 is required for replication in vivo. J. Virol. 2024, 99, e0142324. [Google Scholar] [CrossRef]
- Fradet-Turcotte, A.; Morin, G.; Lehoux, M.; Bullock, P.A.; Archambault, J. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 2010, 399, 65–76. [Google Scholar] [CrossRef]
- Wang, W.; Spurgeon, M.E.; Pope, A.; McGregor, S.; Ward-Shaw, E.; Gronski, E.; Lambert, P.F. Stress keratin 17 and estrogen support viral persistence and modulate the immune environment during cervicovaginal murine papillomavirus infection. Proc. Natl. Acad. Sci. USA 2023, 120, e2214225120. [Google Scholar] [CrossRef]
- Cladel, N.M.; Hu, J.; Balogh, K.; Mejia, A.; Christensen, N.D. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection. J. Virol. Methods 2008, 148, 34–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Kwak, K.; Roden, R.B.; Lowy, D.R.; Schiller, J.T. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology 2012, 433, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Porter, S.S.; McBride, A.A. Human Papillomavirus Quasivirus Production and Infection of Primary Human Keratinocytes. Curr. Protoc. Microbiol. 2020, 57, e101. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-TASSER-MTD: A deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef]
- Sanders, C.M.; Sizov, D.; Seavers, P.R.; Ortiz-Lombardia, M.; Antson, A.A. Transcription activator structure reveals redox control of a replication initiation reaction. Nucleic Acids Res. 2007, 35, 3504–3515. [Google Scholar] [CrossRef]
- Culleton, S.P.; Kanginakudru, S.; DeSmet, M.; Gilson, T.; Xie, F.; Wu, S.Y.; Chiang, C.M.; Qi, G.; Wang, M.; Androphy, E.J. Phosphorylation of the Bovine Papillomavirus E2 Protein on Tyrosine Regulates Its Transcription and Replication Functions. J. Virol. 2017, 91, e01854-16. [Google Scholar] [CrossRef]
- Gilson, T.; Culleton, S.; Xie, F.; DeSmet, M.; Androphy, E.J. Human Papillomavirus 31 Tyrosine 102 Regulates Interaction with E2 Binding Partners and Episomal Maintenance. J. Virol. 2020, 94, e00590-20. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, W.; Gou, Y.; Xu, D.; Wei, Y.; Liu, D.; Han, C.; Huang, X.; Li, C.; Ning, W.; et al. GPS 6.0: An updated server for prediction of kinase-specific phosphorylation sites in proteins. Nucleic Acids Res. 2023, 51, W243–W250. [Google Scholar] [CrossRef]
- Wang, D.; Liang, Y.; Xu, D. Capsule network for protein post-translational modification site prediction. Bioinformatics 2019, 35, 2386–2394. [Google Scholar] [CrossRef]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef]
- Wang, D.; Zeng, S.; Xu, C.; Qiu, W.; Liang, Y.; Joshi, T.; Xu, D. MusiteDeep: A deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics 2017, 33, 3909–3916. [Google Scholar] [CrossRef]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef]
- Dou, Y.; Yao, B.; Zhang, C. PhosphoSVM: Prediction of phosphorylation sites by integrating various protein sequence attributes with a support vector machine. Amino Acids 2014, 46, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cole, P.A. Synthetic approaches to protein phosphorylation. Curr. Opin. Chem. Biol. 2015, 28, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.A.; Naidu, S.R.; Wang, X.; Imbalzano, A.N.; Androphy, E.J. Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation. J. Virol. 2007, 81, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Ustav, M.; Stenlund, A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991, 10, 449–457. [Google Scholar] [CrossRef]
- Sedman, J.; Stenlund, A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995, 14, 6218–6228. [Google Scholar] [CrossRef]
- Skiadopoulos, M.H.; McBride, A.A. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 1996, 70, 1117–1124. [Google Scholar] [CrossRef]
- You, J.; Croyle, J.L.; Nishimura, A.; Ozato, K.; Howley, P.M. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 2004, 117, 349–360. [Google Scholar] [CrossRef]
- Blitz, I.L.; Laimins, L.A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J. Virol. 1991, 65, 649–656. [Google Scholar] [CrossRef]
- Seo, Y.S.; Muller, F.; Lusky, M.; Gibbs, E.; Kim, H.Y.; Phillips, B.; Hurwitz, J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 1993, 90, 2865–2869. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.; DeSmet, M.; Androphy, E.J. A Conserved Di-Lysine Motif in the E2 Transactivation Domain Regulates MmuPV1 Replication and Disease Progression. Pathogens 2025, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Stubenrauch, F.; Straub, E.; Klein, K.; Kramer, D.; Iftner, T.; Wong, M.; Roden, R.B.S. Expression of E8^E2 Is Required for Wart Formation by Mouse Papillomavirus 1 in Vivo. J. Virol. 2021, 95, e01930-20. [Google Scholar] [CrossRef] [PubMed]
- Rank, N.M.; Lambert, P.F. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 1995, 69, 6323–6334. [Google Scholar] [CrossRef]
- Yao, J.M.; Breiding, D.E.; Androphy, E.J. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 1998, 72, 1013–1019. [Google Scholar] [CrossRef]
- Wang, X.; Naidu, S.R.; Sverdrup, F.; Androphy, E.J. Tax1BP1 interacts with papillomavirus E2 and regulates E2-dependent transcription and stability. J. Virol. 2009, 83, 2274–2284. [Google Scholar] [CrossRef]
- Breiding, D.E.; Sverdrup, F.; Grossel, M.J.; Moscufo, N.; Boonchai, W.; Androphy, E.J. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell Biol. 1997, 17, 7208–7219. [Google Scholar] [CrossRef]
- Feeney, K.M.; Saade, A.; Okrasa, K.; Parish, J.L. In vivo analysis of the cell cycle dependent association of the bovine papillomavirus E2 protein and ChlR1. Virology 2011, 414, 1–9. [Google Scholar] [CrossRef]
- Boner, W.; Taylor, E.R.; Tsirimonaki, E.; Yamane, K.; Campo, M.S.; Morgan, I.M. A Functional interaction between the human papillomavirus 16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J. Biol. Chem. 2002, 277, 22297–22303. [Google Scholar] [CrossRef]
- Kasukawa, H.; Howley, P.M.; Benson, J.D. A fifteen-amino-acid peptide inhibits human papillomavirus E1-E2 interaction and human papillomavirus DNA replication in vitro. J. Virol. 1998, 72, 8166–8173. [Google Scholar] [CrossRef] [PubMed]
- Spalholz, B.A.; McBride, A.A.; Sarafi, T.; Quintero, J. Binding of bovine papillomavirus E1 to the origin is not sufficient for DNA replication. Virology 1993, 193, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Mohr, I.J.; Clark, R.; Sun, S.; Androphy, E.J.; MacPherson, P.; Botchan, M.R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 1990, 250, 1694–1699. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Stenlund, A. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 1997, 71, 3853–3863. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Gegonne, A.; Singer, D.S. Bromodomain 4: A cellular Swiss army knife. J. Leukoc. Biol. 2016, 100, 679–686. [Google Scholar] [CrossRef]
- Ilves, I.; Maemets, K.; Silla, T.; Janikson, K.; Ustav, M. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 2006, 80, 3660–3665. [Google Scholar] [CrossRef]
- Lee, A.Y.; Chiang, C.M. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 2009, 284, 2778–2786. [Google Scholar] [CrossRef]
- Schweiger, M.R.; You, J.; Howley, P.M. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 2006, 80, 4276–4285. [Google Scholar] [CrossRef]
- Gagnon, D.; Joubert, S.; Senechal, H.; Fradet-Turcotte, A.; Torre, S.; Archambault, J. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of bromodomain-containing protein 4. J. Virol. 2009, 83, 4127–4139. [Google Scholar] [CrossRef]
- Zheng, G.; Schweiger, M.R.; Martinez-Noel, G.; Zheng, L.; Smith, J.A.; Harper, J.W.; Howley, P.M. Brd4 regulation of papillomavirus protein E2 stability. J. Virol. 2009, 83, 8683–8692. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Meyers, J.M.; Uberoi, A.; Grace, M.; Lambert, P.F.; Munger, K. Cutaneous HPV8 and MmuPV1 E6 Proteins Target the NOTCH and TGF-β Tumor Suppressors to Inhibit Differentiation and Sustain Keratinocyte Proliferation. PLoS Pathog. 2017, 13, e1006171. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Grace, M.; Uberoi, A.; Romero-Masters, J.C.; Lee, D.; Lambert, P.F.; Munger, K. The Mus musculus Papillomavirus Type 1 E7 Protein Binds to the Retinoblastoma Tumor Suppressor: Implications for Viral Pathogenesis. mBio 2021, 12, e0227721. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, J.; DeSmet, M.; Stoll, K.; Jose, L.; Christensen, N.; Androphy, E.J. E2 Tyrosine 102 Regulates MmuPV1 Pathogenesis In Vivo. Pathogens 2025, 14, 913. https://doi.org/10.3390/pathogens14090913
Gonzalez J, DeSmet M, Stoll K, Jose L, Christensen N, Androphy EJ. E2 Tyrosine 102 Regulates MmuPV1 Pathogenesis In Vivo. Pathogens. 2025; 14(9):913. https://doi.org/10.3390/pathogens14090913
Chicago/Turabian StyleGonzalez, Jessica, Marsha DeSmet, Kennedy Stoll, Leny Jose, Neil Christensen, and Elliot J. Androphy. 2025. "E2 Tyrosine 102 Regulates MmuPV1 Pathogenesis In Vivo" Pathogens 14, no. 9: 913. https://doi.org/10.3390/pathogens14090913
APA StyleGonzalez, J., DeSmet, M., Stoll, K., Jose, L., Christensen, N., & Androphy, E. J. (2025). E2 Tyrosine 102 Regulates MmuPV1 Pathogenesis In Vivo. Pathogens, 14(9), 913. https://doi.org/10.3390/pathogens14090913






