Reverse Transcription Recombinase-Aided Amplification Assay for Newcastle Disease Virus in Poultry
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of NDV RT-RAA
2.1.1. Reference Strain
2.1.2. Clinical Samples
2.1.3. RNA Extraction
2.1.4. NDV RT-RAA Primers and Exo-Probe
2.1.5. NDV RT-RAA
2.1.6. Real-Time RT-PCR
2.2. Validation of NDV RT-RAA
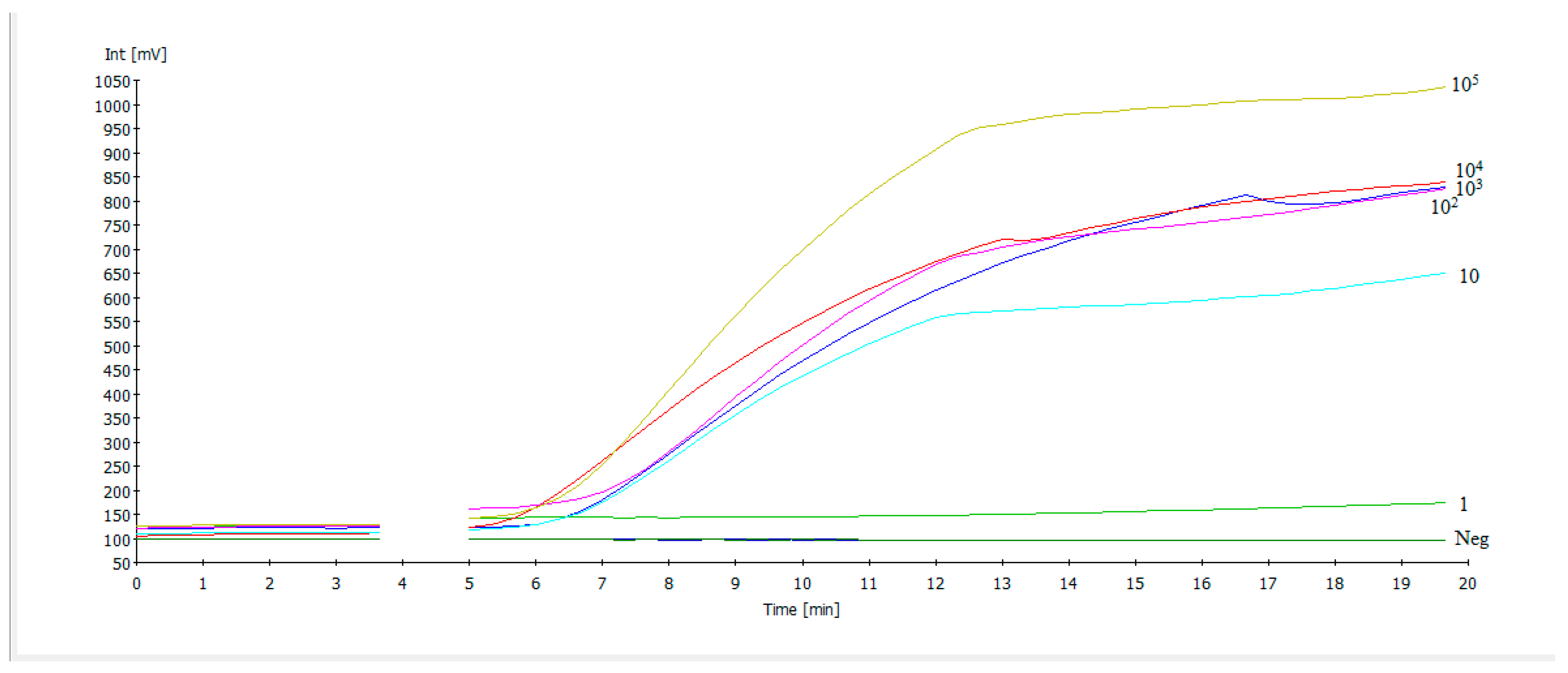
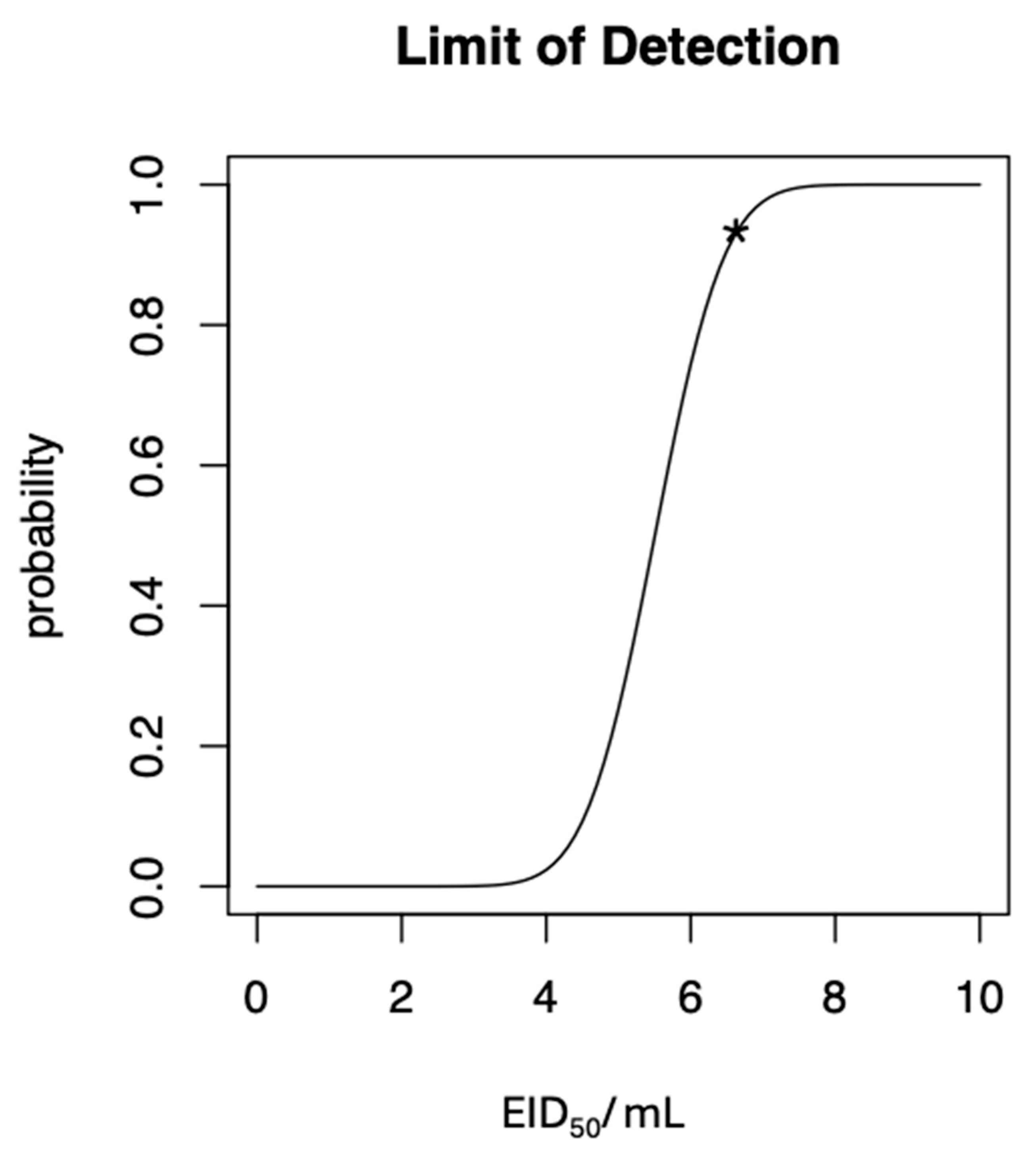
2.2.1. NDV RT-RAA Sensitivity
2.2.2. NDV RT-RAA Specificity and Cross-Reactivity
2.2.3. Clinical Performance of NDV RT-RAA
2.3. Statistical Methods
3. Results
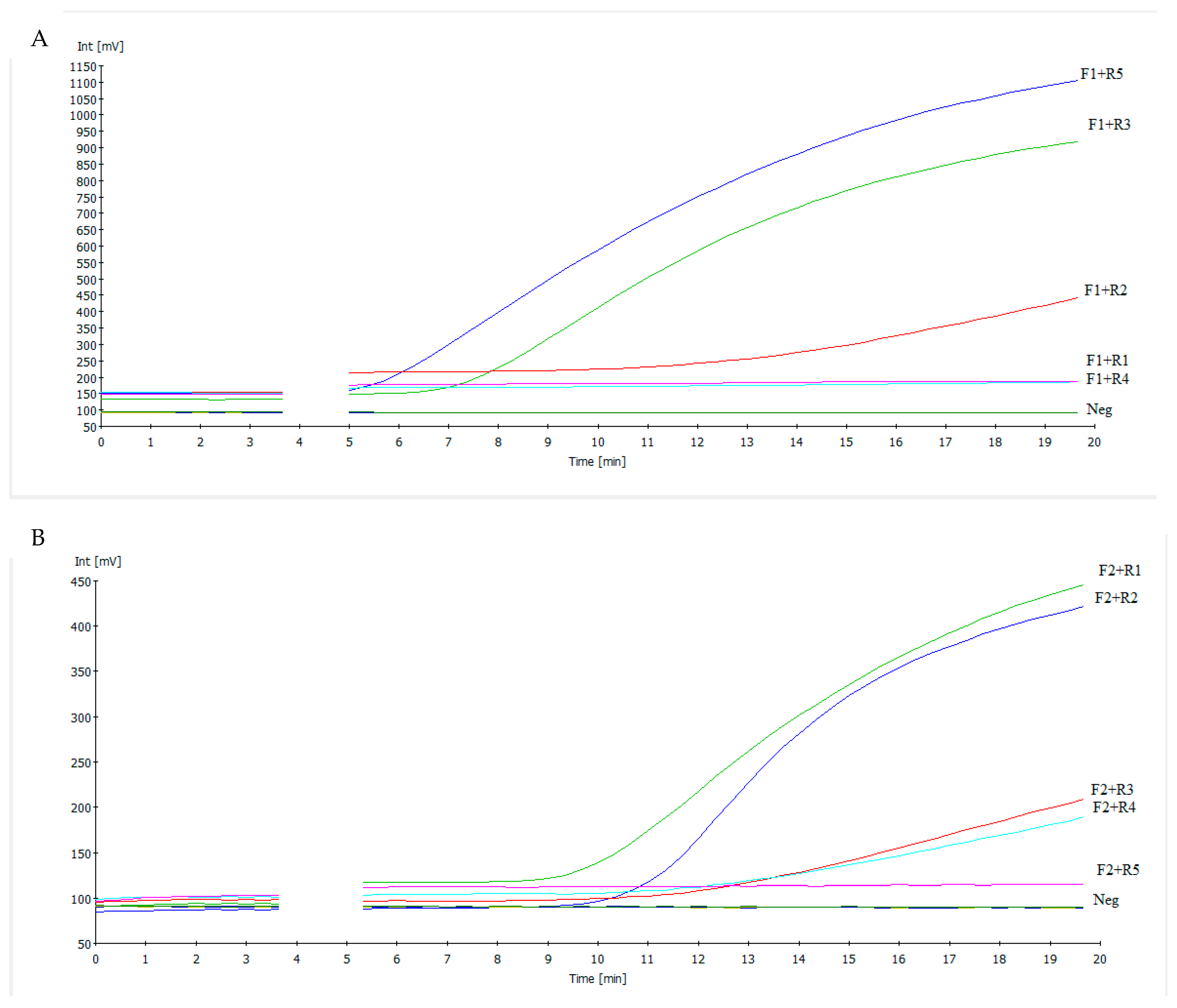
3.1. Primer Selection
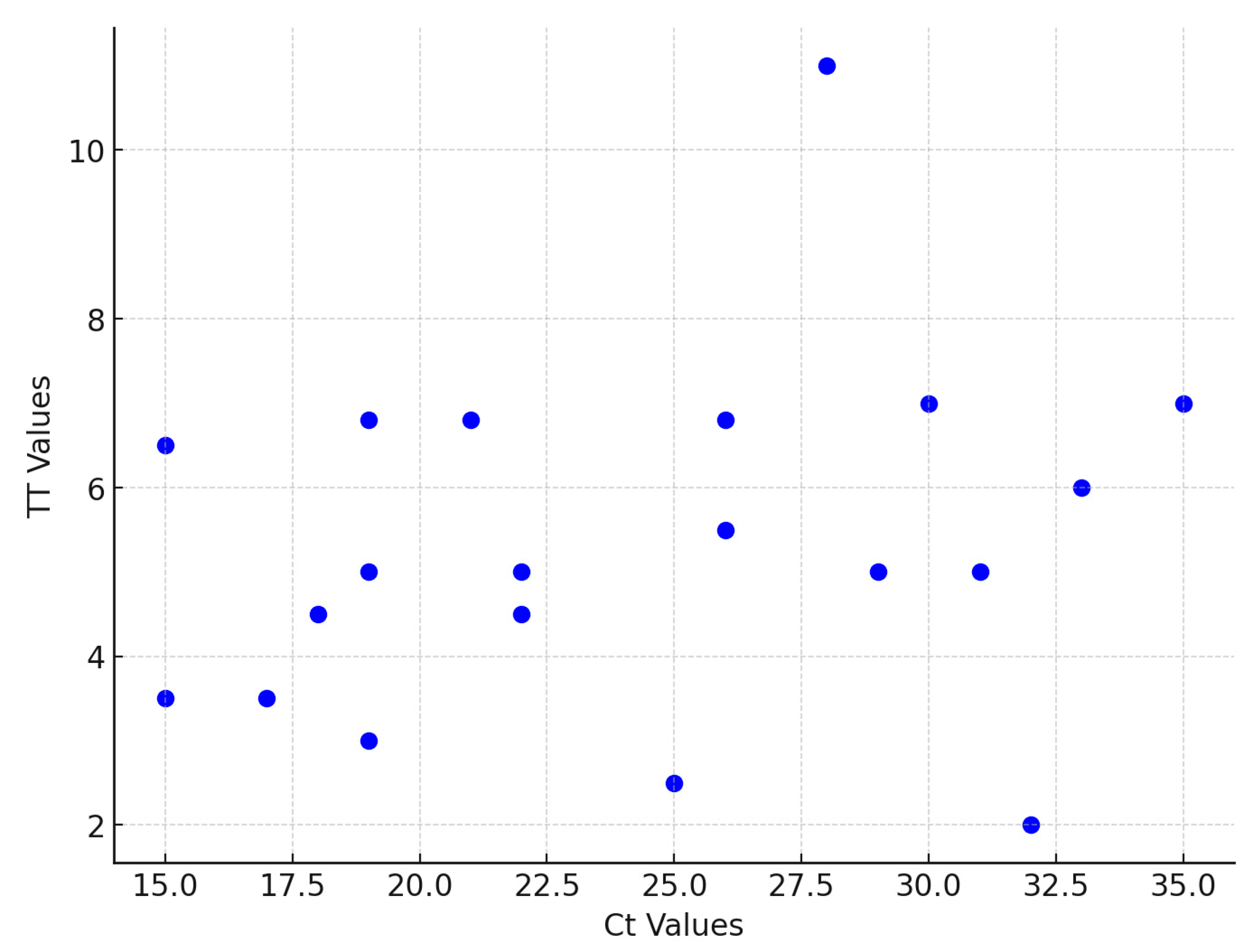
3.2. NDV RT-RAA Assay Sensitivity and Specificity
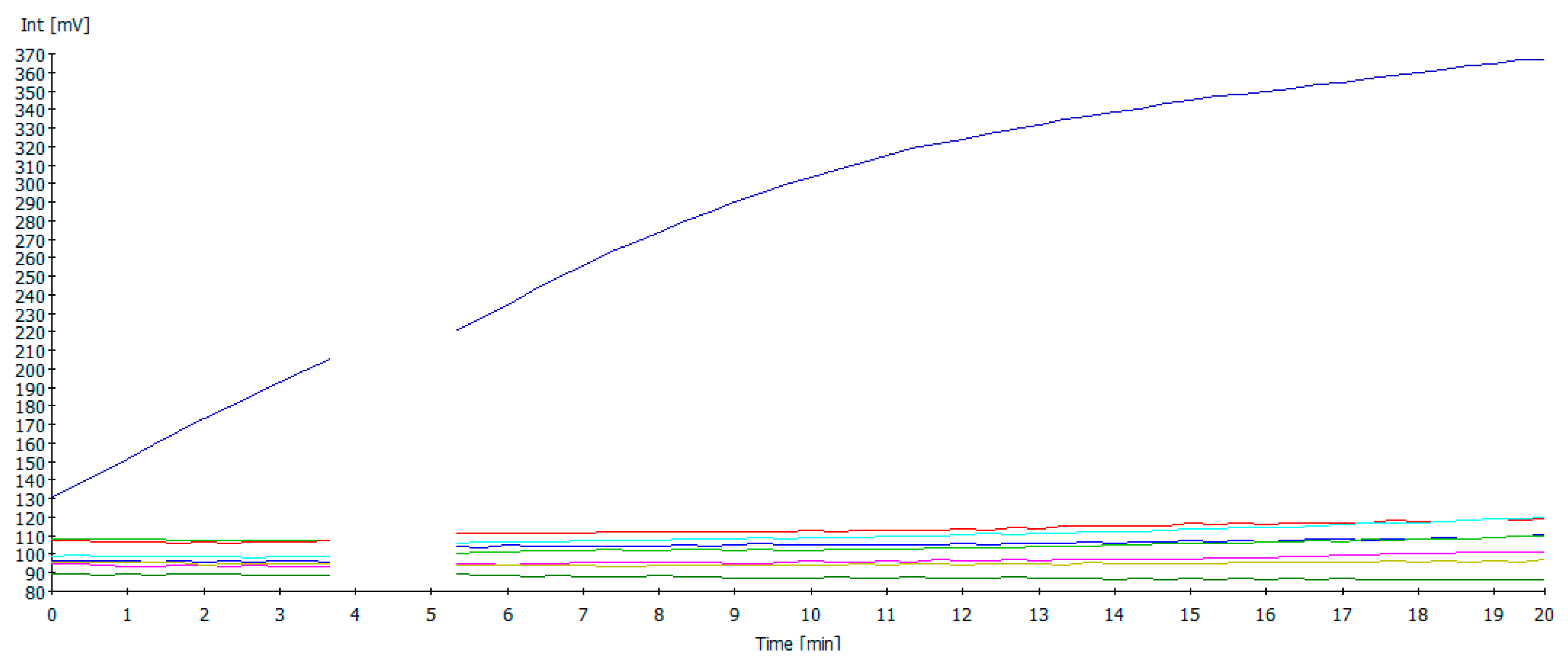
3.3. NDV RT-RAA Cross-Reactivity
3.4. Clinical Performance of NDV RT-RAA Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APMV-1 | Avian paramyxovirus type 1 |
| AOaV-1 | Avian Orthoavulavirus 1 |
| BHQ | Black Hole Quencher |
| CI | Confidence interval |
| CT | Cycle threshold |
| EID50 | 50% embryo infective dose |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| F | Fusion protein |
| FAM | Fluorescein Amidite |
| H5N1 | Highly Pathogenic Avian Influenza subtype H5N1 |
| H7N1 | Highly Pathogenic Avian Influenza subtype H7N1 |
| H9N2 | Low Pathogenic Avian Influenza subtype H9N2 |
| HN | Haemagglutinin-Neuraminidase |
| IBV | Infectious bronchitis virus |
| ILTV | Infectious laryngotracheitis virus |
| L | Large RNA polymerase |
| M | Matrix protein |
| mV | Millivolts |
| N | Nucleocapsid protein |
| ND | Newcastle disease |
| NDV | Newcastle Disease Virus |
| P | Phosphoprotein |
| PBS | Phosphate-buffered saline |
| RAA | Recombinase-aided amplification |
| RPA | Recombinase polymerase amplification |
| RT-PCR | Reverse transcription polymerase chain reaction |
| RT-RAA | Reverse transcription recombinase-aided amplification |
| THF | Tetrahydrofuran |
| TT | Time threshold |
References
- Moustapha, A.; Talaki, E.; Akourki, A.; Ousseini, M. Newcastle disease virus in poultry: Current status and control prospects. World’s Vet. J. 2023, 13, 240–249. [Google Scholar] [CrossRef]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P. ICTV virus taxonomy profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- Diel, D.G.; da Silva, L.H.; Liu, H.; Wang, Z.; Miller, P.J.; Afonso, C.L. Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 2012, 12, 1770–1779. [Google Scholar] [CrossRef]
- Dimitrov, K.M.; Abolnik, C.; Afonso, C.L.; Albina, E.; Bahl, J.; Berg, M.; Briand, F.-X.; Brown, I.H.; Choi, K.-S.; Chvala, I. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019, 74, 103917. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Wu, S.; Hu, S.; Peng, Y.; Xue, F.; Liu, X. Surveillance for avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated. Avian Pathol. 2009, 38, 377–391. [Google Scholar] [CrossRef]
- Aldous, E.; Alexander, D. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. 2001, 30, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Daubney, R.; Mansy, W. The occurrence of Newcastle disease in Egypt. J. Comp. Pathol. 1948, 58, 189–200. [Google Scholar] [CrossRef]
- Abd El-Hamid, H.S.; Shafi, M.E.; Albaqami, N.M.; Ellakany, H.F.; Abdelaziz, N.M.; Abdelaziz, M.N.; Abd El-Hack, M.E.; Taha, A.E.; Alanazi, K.M.; Elbestawy, A.R. Sequence analysis and pathogenicity of Avian Orthoavulavirus 1 strains isolated from poultry flocks during 2015–2019. BMC Vet. Res. 2020, 16, 253. [Google Scholar] [CrossRef]
- El-Bagoury, G.F.; El-Habbaa, A.S.; El-Adaway, S.F.; El-Mahdy, S.S. Isolation, identification and pathotyping of Newcastle disease virus from chickens in Egypt. Benha Vet. Med. J. 2015, 29, 196–204. [Google Scholar] [CrossRef]
- Megahed, M.M.; Eid, A.A.; Mohamed, W.; Hassanin, O. Genetic characterization of Egyptian Newcastle disease virus strains isolated from flocks vaccinated against Newcastle disease virus, 2014–2015. Slov. Vet. Res. 2018, 55, 17–29. [Google Scholar] [CrossRef]
- Moharam, I.; Razik, A.A.e.; Sultan, H.; Ghezlan, M.; Meseko, C.; Franzke, K.; Harder, T.; Beer, M.; Grund, C. Investigation of suspected Newcastle disease (ND) outbreaks in Egypt uncovers a high virus velogenic ND virus burden in small-scale holdings and the presence of multiple pathogens. Avian Pathol. 2019, 48, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Chumbe, A.; Izquierdo-Lara, R.; Calderón, K.; Fernández-Díaz, M.; Vakharia, V.N. Development of a novel Newcastle disease virus (NDV) neutralization test based on recombinant NDV expressing enhanced green fluorescent protein. Virol. J. 2017, 14, 232. [Google Scholar] [CrossRef]
- Franzo, G.; Massi, P.; Tucciarone, C.M.; Barbieri, I.; Tosi, G.; Fiorentini, L.; Ciccozzi, M.; Lavazza, A.; Cecchinato, M.; Moreno, A. Think globally, act locally: Phylodynamic reconstruction of infectious bronchitis virus (IBV) QX genotype (GI-19 lineage) reveals different population dynamics and spreading patterns when evaluated on different epidemiological scales. PLoS ONE 2017, 12, e0184401. [Google Scholar] [CrossRef]
- Yehia, N.; Salem, H.M.; Mahmmod, Y.; Said, D.; Samir, M.; Mawgod, S.A.; Sorour, H.K.; AbdelRahman, M.A.; Selim, S.; Saad, A.M. Common viral and bacterial avian respiratory infections: An updated review. Poult. Sci. 2023, 102, 102553. [Google Scholar] [CrossRef]
- Mao, Q.; Ma, S.; Schrickel, P.L.; Zhao, P.; Wang, J.; Zhang, Y.; Li, S.; Wang, C. Review detection of Newcastle disease virus. Front. Vet. Sci. 2022, 9, 936251. [Google Scholar] [CrossRef]
- Bello, M.B.; Yusoff, K.; Ideris, A.; Hair-Bejo, M.; Peeters, B.P.; Omar, A.R. Diagnostic and vaccination approaches for Newcastle disease virus in poultry: The current and emerging perspectives. BioMed Res. Int. 2018, 2018, 7278459. [Google Scholar] [CrossRef]
- Wise, M.G.; Suarez, D.L.; Seal, B.S.; Pedersen, J.C.; Senne, D.A.; King, D.J.; Kapczynski, D.R.; Spackman, E. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004, 42, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, P.P.; Zhang, Y.H.; Tian, K.Y.; Bian, C.Z.; Zhao, J. Development of a reverse transcription recombinase polymerase amplification combined with lateral–flow dipstick assay for avian influenza H9N2 HA gene detection. Transbound. Emerg. Dis. 2019, 66, 546–551. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Wang, G.; Zhang, Y.; Shang, Y.; Zhang, Z. Development of a fluorescent probe-based recombinase polymerase amplification assay for rapid detection of Orf virus. Virol. J. 2015, 12, 206. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Zhang, W.; Li, Z.; Zhang, S.; Li, Y.; Zhang, Z. Development of an isothermal recombinase polymerase amplification assay for rapid detection of pseudorabies virus. Mol. Cell. Probes 2017, 33, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Yehia, N.; Arafa, A.-S.; Abd El Wahed, A.; El-Sanousi, A.A.; Weidmann, M.; Shalaby, M.A. Development of reverse transcription recombinase polymerase amplification assay for avian influenza H5N1 HA gene detection. J. Virol. Methods 2015, 223, 45–49. [Google Scholar] [CrossRef]
- Yehia, N.; Eldemery, F.; Arafa, A.-S.; Abd El Wahed, A.; El Sanousi, A.; Weidmann, M.; Shalaby, M. Reverse transcription recombinase polymerase amplification assay for rapid detection of avian influenza virus H9N2 HA gene. Vet. Sci. 2021, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zeng, F.; Sun, J.; Liu, X.; Wu, M.; Huang, B.; Lian, Y.; Xiao, L.; Ma, L.; Zhang, S. Application of recombinase polymerase amplification method for rapid detection of infectious laryngotracheitis virus. Mol. Cell. Probes 2020, 54, 101646. [Google Scholar] [CrossRef]
- Lomniczi, B.; Wehmann, E.; Herczeg, J.; Ballagi-Pordany, A.; Kaleta, E.; Werner, O.; Meulemans, G.; Jorgensen, P.; Mante, A.; Gielkens, A. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII). Arch. Virol. 1998, 143, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Seal, B.S.; King, D.J.; Locke, D.P.; Senne, D.A.; Jackwood, M.W. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 1998, 36, 1141–1145. [Google Scholar] [CrossRef]
- Seal, B.S.; King, D.J.; Meinersmann, R.J. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res. 2000, 66, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kishida, N.; Sakoda, Y.; Eto, M.; Sunaga, Y.; Kida, H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch. Virol. 2004, 149, 2095–2104. [Google Scholar] [CrossRef]
- Parikh, R.; Mathai, A.; Parikh, S.; Sekhar, G.C.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- Alexander, D.J. Newcastle disease. Br. Poult. Sci. 2001, 42, 5–22. [Google Scholar] [CrossRef]
- Jang, J.; Hong, S.-H.; Kim, I.-H. Validation of a real-time RT-PCR method to quantify Newcastle disease virus (NDV) titer and comparison with other quantifiable methods. J. Microbiol. Biotechnol. 2011, 21, 100–108. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, D.; Hu, J.; Sun, W.; Liu, K.; Li, J.; Xu, H.; Liu, J.; He, L.; Jiang, D. Multiplex one-step real-time PCR assay for rapid simultaneous detection of velogenic and mesogenic Newcastle disease virus and H5-subtype avian influenza virus. Arch. Virol. 2019, 164, 1111–1119. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, C.; Bai, Y.; Zhang, P.; Yao, S.; Liu, J.; Zhang, T. Establishment of reverse transcription recombinase–aided amplification-lateral-flow dipstick and real-time fluorescence–based reverse transcription recombinase–aided amplification methods for detection of the Newcastle disease virus in chickens. Poult. Sci. 2020, 99, 3393–3401. [Google Scholar] [CrossRef] [PubMed]
- Ganar, K.; Das, M.; Sinha, S.; Kumar, S. Newcastle disease virus: Current status and our understanding. Virus Res. 2014, 184, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-S.; Kim, H.-S.; Kim, J.-Y.; Kwon, Y.-K.; Kim, H.-R. The Development of Novel Reverse Transcription Loop-Mediated Isothermal Amplification Assays for the Detection and Differentiation of Virulent Newcastle Disease Virus. Int. J. Mol. Sci. 2023, 24, 13847. [Google Scholar] [CrossRef] [PubMed]
- Gunaratna, G.; Manamperi, A.; Böhlken-Fascher, S.; Wickremasinge, R.; Gunawardena, K.; Yapa, B.; Pathirana, N.; Pathirana, H.; de Silva, N.; Sooriyaarachchi, M. Evaluation of rapid extraction and isothermal amplification techniques for the detection of Leishmania donovani DNA from skin lesions of suspected cases at the point of need in Sri Lanka. Parasites Vectors 2018, 11, 665. [Google Scholar] [CrossRef]
- Schurig, S.; Ceruti, A.; Wende, A.; Lübcke, P.; Eger, E.; Schaufler, K.; Frimpong, M.; Truyen, U.; Kobialka, R.M.; Abd El Wahed, A. Rapid Identification of Bacterial Composition in Wastewater by Combining Reverse Purification Nucleic Acid Extraction and Nanopore Sequencing. ACS EST Water 2024, 4, 1808–1818. [Google Scholar] [CrossRef]
| Name | Sequence |
|---|---|
| NDV-F1 | 5′AGAAAGTGACATTTGACAAGATAGAGGGAAAG 3′ |
| NDV-F2 | 5′AGTGACATTTGACAAGATAGAGGGAAAGATAAG3′ |
| NDV-R1 | 5′AGAGGCATTTGCTATAGGATAGCAGGCCGTC3′ |
| NDV-R2 | 5′CTTATCTTTCCCTCTATCTTGTCAAATGTCACT3′ |
| NDV-R3 | 5′CCTGAGGGGAGGCATTTGCTATAGGATAGCAG3′ |
| NDV-R4 | 5′CAACCTGAGGGGAGGCATTTGCTATAGGA3′ |
| NDV-R5 | 5′CCTGGGGAGAGGCATTTGCTATAGGATAG3′ |
| NDV-exo-probe1 | 5′GCTCAGTGATGTGCTCGGACCCTCTG(BHQ1-dT)(THF) C (FAM-dT)TGTGAAGGCGAGAG-PH 3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yehia, N.; Abd El Wahed, A.; Mohamed, A.A.E.; Arafa, A.; Said, D.; Shalaby, M.A.; Ceruti, A.; Truyen, U.; Kobialka, R.M. Reverse Transcription Recombinase-Aided Amplification Assay for Newcastle Disease Virus in Poultry. Pathogens 2025, 14, 867. https://doi.org/10.3390/pathogens14090867
Yehia N, Abd El Wahed A, Mohamed AAE, Arafa A, Said D, Shalaby MA, Ceruti A, Truyen U, Kobialka RM. Reverse Transcription Recombinase-Aided Amplification Assay for Newcastle Disease Virus in Poultry. Pathogens. 2025; 14(9):867. https://doi.org/10.3390/pathogens14090867
Chicago/Turabian StyleYehia, Nahed, Ahmed Abd El Wahed, Ahmed Abd Elhalem Mohamed, Abdelsattar Arafa, Dalia Said, Mohamed A. Shalaby, Arianna Ceruti, Uwe Truyen, and Rea Maja Kobialka. 2025. "Reverse Transcription Recombinase-Aided Amplification Assay for Newcastle Disease Virus in Poultry" Pathogens 14, no. 9: 867. https://doi.org/10.3390/pathogens14090867
APA StyleYehia, N., Abd El Wahed, A., Mohamed, A. A. E., Arafa, A., Said, D., Shalaby, M. A., Ceruti, A., Truyen, U., & Kobialka, R. M. (2025). Reverse Transcription Recombinase-Aided Amplification Assay for Newcastle Disease Virus in Poultry. Pathogens, 14(9), 867. https://doi.org/10.3390/pathogens14090867








