Adaptation and Outbreak of Highly Pathogenic Avian Influenza in Dairy Cattle: An Emerging Threat to Humans, Pets, and Peridomestic Animals
Abstract
1. Introduction
2. Literature Search
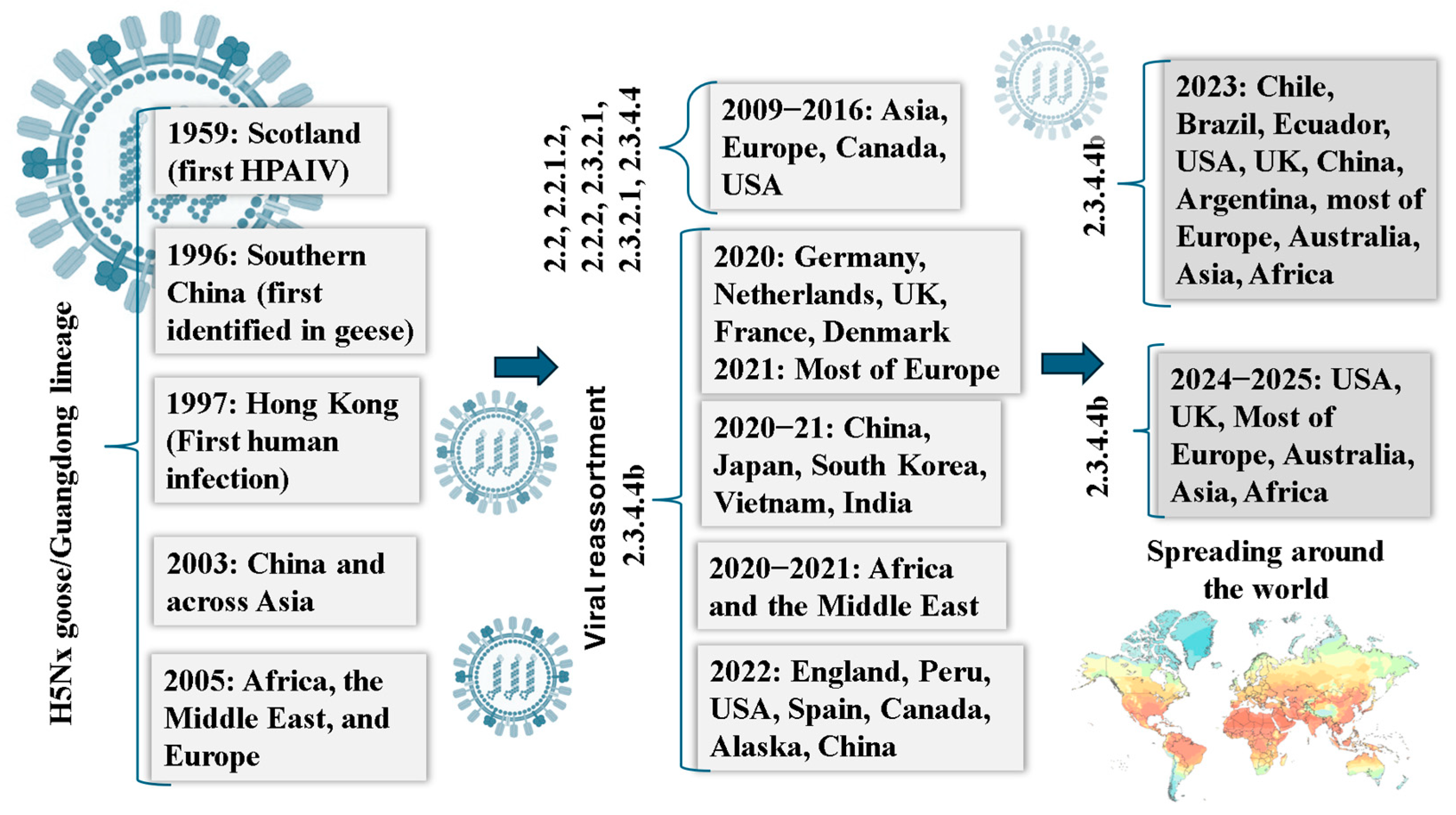
3. Influenza A Virus: Structure, Ecology, and the Threat of Host Expansion
4. Unprecedented Emergence of Influenza in Bovine Hosts
5. From Birds to Mammals: Cross-Species Spillover of HPAI H5N1 with Emphasis on U.S. Dairy Herds
5.1. Transmission Pathways and Interspecies Spread of HPAI H5N1 Clade 2.3.4.4b in U.S. Dairy Cattle
5.2. Rethinking Tropism: The Mammary Gland Plays a Critical Role in Virus Evolution and Transmission
5.3. Emerging Threat of HPAI H5N1 in Mammals with a Focus on Recent Feline Infections Linked to Dairy Cattle
6. H5N1 in the Dairy Industry: Zoonotic Spillover and Impact on Public Health
7. Occupational Exposure: A Significant Pathway for Virus Spillover to and from Humans
8. Recent Development of Immunization Strategies for HPAI H5N1 in Dairy Cattle
9. Strengthening Pandemic Preparedness: Lessons from the HPAI H5N1 Outbreaks in Dairy Cattle and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burrough, E.R.; Magstadt, D.R.; Petersen, B.; Timmermans, S.J.; Gauger, P.C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A.C.; et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerg. Infect. Dis. 2024, 30, 1335–1343. [Google Scholar] [CrossRef]
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic potential of influenza A viruses: A comprehensive overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Naguib, M.M.; Nogales, A.; Barre, R.S.; Stewart, J.P.; García-Sastre, A.; Martinez-Sobrido, L. Avian influenza A (H5N1) virus in dairy cattle: Origin, evolution, and cross-species transmission. Am. Soc. Microbiol. 2024, 15, e0254224. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Thomas, M.; Kaushik, R.S.; Wang, D.; Li, F. Influenza A in Bovine Species: A Narrative Literature Review. Viruses 2019, 11, 561. [Google Scholar] [CrossRef]
- Liu, R.; Sheng, Z.; Huang, C.; Wang, D.; Li, F. Influenza D virus. Curr. Opin. Virol. 2020, 44, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, Y.; Katsushima, N.; Nagai, Y.; Shoji, M.; Itagaki, T.; Sakamoto, M.; Nishimura, H. Clinical features of influenza C virus infection in children. J. Infect. Dis. 2006, 193, 1229–1235. [Google Scholar] [CrossRef]
- Kaplan, B.S.; Falkenberg, S.; Dassanayake, R.; Neill, J.; Velayudhan, B.; Li, F.; Vincent, A.L. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound. Emerg. Dis. 2021, 68, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Martinez-Sobrido, L.; Mostafa, A. Zoonosis and zooanthroponosis of emerging respiratory viruses. Front. Cell. Infect. Microbiol. 2024, 13, 1232772. [Google Scholar] [CrossRef]
- Youk, S.; Torchetti, M.K.; Lantz, K.; Lenoch, J.B.; Killian, M.L.; Leyson, C.; Pantin-Jackwood, M.J. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021–April 2022. Virology 2023, 587, 109860. [Google Scholar] [CrossRef]
- Bevins, S.N.; Shriner, S.A.; Cumbee Jr, J.C.; Dilione, K.E.; Douglass, K.E.; Ellis, J.W.; Lenoch, J.B. Intercontinental Movement of Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4 Virus to the United States, 2021. Emerg. Infect. Dis. 2022, 28, 1006–1011. [Google Scholar] [CrossRef]
- Hu, X.; Saxena, A.; Magstadt, D.R.; Gauger, P.C.; Burrough, E.; Zhang, J.; Siepker, C.; Mainenti, M.; Gorden, P.J.; Plummer, P.; et al. Highly Pathogenic Avian Influenza A (H5N1) clade 2.3.4.4b Virus detected in dairy cattle. bioRxiv, 2024; bioRxiv:2024.04.16.588916. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Hutter, C.R.; Markin, A.; Thomas, M.; Lantz, K.; Killian, M.L.; Janzen, G.M.; Vijendran, S.; Wagle, S.; Inderski, B.; et al. Emergence and interstate spread of highly pathogenic avian influenza A(H5N1) in dairy cattle in the United States. Science 2025, 388, eadq0900. [Google Scholar] [CrossRef]
- Mitchell, C.A.; Walker, R.V.; Bannister, G.L. Further Experiments Relating to the Propagation of Virus in the Bovine Mammary Gland. Can. J. Comp. Med. Vet. Sci. 1953, 17, 218–222. [Google Scholar]
- Mitchell, C.A.; Walker, R.V.; Bannister, G.L. Studies relating to the formation of neutralizing antibodies following the propagation of influenza and Newcastle disease virus in the bovine mammary gland. Can. J. Microbiol. 1956, 2, 322–328. [Google Scholar] [CrossRef] [PubMed]
- CIRDAP Report, Nevada Reports H5N1 in Dairy Worker; USDA Fleshes out D1.1 Sequencing from Affected Herds. 2025. Available online: https://www.cidrap.umn.edu/nevada-reports-h5n1-dairy-worker-usda-fleshes-out-d11-sequencing-affected-herds (accessed on 12 February 2025).
- Uyeki, T.M.; Milton, S.; Abdul Hamid, C.; Reinoso Webb, C.; Presley, S.M.; Shetty, V.; Rollo, S.N.; Martinez, D.L.; Rai, S.; Gonzales, E.R.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Infection in a Dairy Farm Worker. N. Engl. J. Med. 2024, 390, 2028–2029. [Google Scholar] [CrossRef]
- Naraharisetti, R. Highly pathogenic avian influenza A (H5N1) virus infection of indoor domestic cats within dairy industry worker households—Michigan, May 2024. MMWR Morb. Mortal. Wkly. Rep. 2025, 74, 61–65. [Google Scholar] [CrossRef]
- Gultyaev, A.P.; Fouchier, R.A.; Olsthoorn, R.C. Influenza virus RNA structure: Unique and common features. Int. Rev. Immunol. 2010, 29, 533–556. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; von Kirchbach, J.C.; Gog, J.R.; Digard, P. Genome packaging in influenza A virus. J. Gen. Virol. 2010, 91 Pt 2, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Ruigrok, R.W.; Crépin, T.; Hart, D.J.; Cusack, S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 2010, 20, 104–113. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Li, F.; Wang, D. Cross-Species Transmission of Highly Pathogenic Avian Influenza (HPAI) H5N1 Virus in the U.S. Dairy Cattle: A Comprehensive Review. Preprints 2024, 2024052137. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.F.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, Y.; Rivailler, P.; Conrady, C.; Castillo, D.A.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, L.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef]
- Yoon, S.W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 2014, 385, 359–375. [Google Scholar] [CrossRef]
- Webster, R.G.; Thomas, T.L. Efficacy of equine influenza vaccines for protection against A/Equine/Jilin/89 (H3N8)—A new equine influenza virus. Vaccine 1993, 11, 987–993. [Google Scholar] [CrossRef]
- De Graaf, M.; Fouchier, R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014, 33, 823–841. [Google Scholar] [CrossRef]
- Zhao, C.; Pu, J. Influence of Host Sialic Acid Receptors Structure on the Host Specificity of Influenza Viruses. Viruses 2022, 14, 2141. [Google Scholar] [CrossRef] [PubMed]
- Kimble, B.; Nieto, G.R.; Perez, D.R. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J. 2010, 7, 365. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the center: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, C.; Han, K.Y.; Zhang, F.X.; Zhu, Y.L.; Ling, Z.S.; Zhang, X.X.; Jiang, S.J.; Xie, Z.J. Molecular characterization of H9N2 influenza virus isolated from mink and its pathogenesis in mink. Vet. Microbiol. 2015, 176, 88–96. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Li, F.; Wang, D. Emerging Threats of Highly Pathogenic Avian Influenza A (H5N1) in US Dairy Cattle: Understanding Cross-Species Transmission Dynamics in Mammalian Hosts. Viruses 2024, 16, 1703. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kamel, M.; Aleya, S.; Almagharbeh, W.T.; Aleya, L.; Abdel-Daim, M.M. The emergence of highly pathogenic avian influenza H5N1 in dairy cattle: Implications for public health, animal health, and pandemic preparedness. Eur. J. Clin. Microbiol. Infect. Dis. 2025, 44, 1817–1833. [Google Scholar] [CrossRef]
- Saito, K. An outbreak of cattle influenza in Japan in the fall of 1949. J. Am. Vet. Med. Assoc. 1951, 118, 316–319. [Google Scholar]
- Romvary, J.; Takatsy, G.; BARB, K.; Farkas, E. Isolation of Influenza Virus Strains from Animals. Nature 1962, 193, 907–908. [Google Scholar] [CrossRef]
- Schild, G.C.; Newman, R.W.; Webster, R.G.; Major, D.; Hinshaw, V.S. Antigenic analysis of influenza A virus surface antigens: Considerations for the nomenclature of influenza virus. Arch. Virol. 1980, 63, 171–184. [Google Scholar] [CrossRef]
- Campbell, C.H.; Easterday, B.C.; Webster, R.G. Strains of Hong Kong influenza virus in calves. J. Infect. Dis. 1977, 135, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Fatkhuddinova, M.F.; Kir’ianova, A.I.; Isachenko, V.A.; Zakstel’skaia, L.I. Vydelenie i identifikatsiia virusa grippa A-Gonkong (H3N2) pri respiratornykh zabolevaniiakh krupnogo rogatogo skota [Isolation and identification of the A-Hong Kong (H3N2) virus in respiratory diseases of cattle]. Vopr. Virusol. 1973, 18, 474–478. [Google Scholar]
- Tanyi, J.; Romváry, J.; Aldásy, P.; Mathé, Z. Isolation of influenza a virus strains from cattle. Preliminary report. Acta Vet. Acad. Sci. Hung. 1974, 24, 341–343. [Google Scholar] [PubMed]
- Brown, I.H.; Crawshaw, T.R.; Harris, P.A.; Alexander, D.J. Detection of antibodies to influenza A virus in cattle in association with respiratory disease and reduced milk yield. Vet. Rec. 1998, 143, 637–638. [Google Scholar] [PubMed]
- Gunning, R.F.; Pritchard, G.C. Unexplained sporadic milk drop in dairy cows. Vet. Rec. 1997, 140, 488. [Google Scholar] [PubMed]
- Crawshaw, T.R.; Brown, I.H.; Essen, S.C.; Young, S.C. Significant rising antibody titers to influenza A are associated with an acute reduction in milk yield in cattle. Vet. J. 2008, 178, 98–102. [Google Scholar] [CrossRef]
- Baker, A.L.; Arruda, B.; Palmer, M.V.; Boggiatto, P.; Sarlo Davila, K.; Buckley, A.; Gorden, P.J. Dairy cows inoculated with highly pathogenic avian influenza virus H5N1. Nature 2024, 637, 913–920. [Google Scholar] [CrossRef]
- Lowen, A.C.; Baker, A.L.; Bowman, A.S.; García-Sastre, A.; Hensley, S.E.; Lakdawala, S.S.; Moncla, L.H.; Nelson, M.I.; Pekosz, A.; Poulson, R.L.; et al. Pandemic risk stemming from the bovine H5N1 outbreak: An account of the knowns and unknowns. J. Virol. 2025, 99, e00052-25. [Google Scholar] [CrossRef]
- Krammer, F.; Schultz-Cherry, S. We need to keep an eye on avian influenza. Nat. Rev. Immunol. 2023, 23, 267–268. [Google Scholar] [CrossRef]
- Kiley, J.L.; Yun, H.C. Avian Influenza: An Overview and Clinical Status. Curr. Infect. Dis. Rep. 2025, 27, 1. [Google Scholar] [CrossRef]
- Dey, P.; Ahuja, A.; Panwar, J.; Choudhary, P.; Rani, S.; Kaur, M.; Sharma, A.; Kaur, J.; Yadav, A.K.; Sood, V.; et al. Immune Control of Avian Influenza Virus Infection and Its Vaccine Development. Vaccines 2023, 11, 593. [Google Scholar] [CrossRef]
- Charostad, J.; Rukerd, M.R.Z.; Mahmoudvand, S.; Bashash, D.; Hashemi, S.M.A.; Nakhaie, M.; Zandi, K. A comprehensive review of highly pathogenic avian influenza (HPAI) H5N1: An imminent threat at doorstep. Travel. Med. Infect. Dis. 2023, 55, 102638. [Google Scholar] [CrossRef]
- Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Mirinavičiūtė, G.; Niqueux, É.; Ståhl, K.; Staubach, C.; Terregino, C.; Willgert, K.; et al. Avian influenza overview September-December 2023. EFSA J. 2023, 21, e8539. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- USDA (U.S. Department of Agriculture). Detections of Highly Pathogenic Avian Influenza (HPAI) in Livestock. Available online: https://www.aphis.usda.gov/livestock-poultry-disease/avian/avianinfluenza/hpai-detections/livestock (accessed on 21 August 2024).
- CDC Report 12th February, 2025, Current Situation: Bird Flu in Dairy Cows. Available online: https://www.cdc.gov/bird-flu/situation-summary/mammals.html (accessed on 12 February 2025).
- Rawson, T.; Morgenstern, C.; Knock, E.S.; Hicks, J.; Pham, A.; Morel, G.; Murillo, A.C.; Sanderson, M.W.; Forchini, G.; FitzJohn, R.; et al. A mathematical model of H5N1 influenza transmission in US dairy cattle. Nat. Commun. 2025, 16, 4308. [Google Scholar] [CrossRef]
- Caserta, L.C.; Frye, E.A.; Butt, S.L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L.M.; Thompson, A.C.; Koscielny, M.P.; Cronk, B.; Johnson, A.; et al. Spillover of highly pathogenic avian influenza H5N1 virus to dairy cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- USDA (U.S. Department of Agriculture). Highly Pathogenic Avian Influenza H5N1 Genotype B3.13 in Dairy Cattle: National Epidemiologic Brief Overview. Available online: https://www.aphis.usda.gov/sites/default/files/hpai-dairy-national-epi-brief (accessed on 8 June 2024).
- Rodriguez, Z.; O’Connor, A.; Bradford, B.J.; Picasso-Risso, C. Characterization and health, productivity, and economic effects of highly pathogenic avian influenza H5N1 outbreak in dairy cattle. J. Dairy. Sci. 2025, 108, 6349–6358. [Google Scholar] [CrossRef] [PubMed]
- Halwe, N.J.; Cool, K.; Breithaupt, A.; Schön, J.; Trujillo, J.D.; Nooruzzaman, M.; Kwon, T.; Ahrens, A.K.; Britzke, T.; McDowell, C.D.; et al. H5N1 clade 2.3.4.4b dynamics in experimentally infected calves and cows. Nature 2024, 637, 903–912. [Google Scholar] [CrossRef]
- Martins, R.P.; Marc, D.; Germon, P.; Trapp, S.; Caballero-Posadas, I. Influenza A virus in dairy cattle: Infection biology and potential mammary gland-targeted vaccines. NPJ Vaccines 2025, 10, 8. [Google Scholar] [CrossRef]
- CDC. Influenza (flu): Technical Report: Highly Pathogenic Avian Influenza A(H5N1) Viruses; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2024. Available online: https://www.cdc.gov/bird-flu/php/technical-report/h5n1-04262024.html (accessed on 26 April 2024).
- Kalthoff, D.; Hoffmann, B.; Harder, T.; Durban, M.; Beer, M. Experimental infection of cattle with highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 2008, 14, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Nelli, R.K.; Harm, T.A.; Siepker, C.; Groeltz-Thrush, J.M.; Jones, B.; Twu, N.C.; Nenninger, A.S.; Magstadt, D.R.; Burrough, E.R.; Piñeyro, P.E.; et al. Sialic Acid Receptor Specificity in Mammary Gland of Dairy Cattle Infected with Highly Pathogenic Avian Influenza A(H5N1) Virus. Emerg. Infect. Dis. 2024, 30, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, C.; Jensen, H.E.; Trebbien, R.; Webby, R.J.; Larsen, L.E. Avian and Human Influenza A Virus Receptors in Bovine Mammary Gland. Emerg. Infect. Dis. 2024, 30, 1907. [Google Scholar] [CrossRef]
- Imai, M.; Ueki, H.; Ito, M.; Iwatsuki-Horimoto, K.; Kiso, M.; Biswas, A.; Trifkovic, S.; Cook, N.; Halfmann, P.J.; Neumann, G.; et al. Highly pathogenic avian H5N1 influenza A virus replication in ex vivo cultures of bovine mammary gland and teat tissues. Emerg. Microbes Infect. 2025, 14, 2450029. [Google Scholar] [CrossRef]
- Gorden, P.J.; Magstadt, D.R.; Baker, A.L.; Arruda, B.L.; Bell, T.M.; Nelli, R.K. Viral Mastitis Associated with Influenza A in Dairy Cattle. Vet. Clin. N. Am. Food Anim. Pract. 2025, 41, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A.J.; Brizuela, K.; Lakdawala, S.S. mGem: Transmission and exposure risks of dairy cow H5N1 influenza virus. mBio 2025, 16, e02944-24. [Google Scholar] [CrossRef]
- Murawski, A.; Fabrizio, T.; Ossiboff, R.; Kackos, C.; Jeevan, T.; Jones, J.C.; Kandeil, A.; Walker, D.; Turner, J.C.; Patton, C.; et al. Highly pathogenic avian influenza A(H5N1) virus in a common bottlenose dolphin (Tursiops truncatus) in Florida. Commun. Biol. 2024, 7, 476. [Google Scholar] [CrossRef]
- Jakobek, B.T.; Berhane, Y.; Nadeau, M.S.; Embury-Hyatt, C.; Lung, O.; Xu, W.; Lair, S. Influenza A(H5N1) Virus Infections in 2 Free-Ranging Black Bears (Ursus americanus), Quebec, Canada. Emerg. Infect. Dis. 2023, 29, 2145–2149. [Google Scholar] [CrossRef]
- Puryear, W.; Sawatzki, K.; Hill, N.; Foss, A.; Stone, J.J.; Doughty, L.; Walk, D.; Gilbert, K.; Murray, M.; Cox, E.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Outbreak in New England Seals, United States. Emerg. Infect. Dis. 2023, 29, 786–791. [Google Scholar] [CrossRef]
- Keawcharoen, J.; Oraveerakul, K.; Kuiken, T.; Fouchier, R.A.; Amonsin, A.; Payungporn, S.; Noppornpanth, S.; Wattanodorn, S.; Theambooniers, A.; Tantilertcharoen, R.; et al. Avian influenza H5N1 in tigers and leopards. Emerg. Infect. Dis. 2004, 10, 2189–2191. [Google Scholar] [CrossRef]
- Agüero, M.; Monne, I.; Sánchez, A.; Zecchin, B.; Fusaro, A.; Ruano, M.J.; Del Valle Arrojo, M.; Fernández-Antonio, R.; Souto, A.M.; Tordable, P.; et al. Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Eurosurveillance 2023, 28, 2300001. [Google Scholar] [CrossRef]
- Elsmo, E.J.; Wünschmann, A.; Beckmen, K.B.; Broughton-Neiswanger, L.E.; Buckles, E.L.; Ellis, J.; Fitzgerald, S.D.; Gerlach, R.; Hawkins, S.; Ip, H.S.; et al. Highly Pathogenic Avian Influenza A(H5N1) Virus Clade 2.3.4.4b Infections in Wild Terrestrial Mammals, United States, 2022. Emerg. Infect. Dis. 2023, 29, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Plaza, P.I.; Gamarra-Toledo, V.; Euguí, J.R.; Rosciano, N.; Lambertucci, S.A. Pacific and Atlantic sea lion mortality caused by highly pathogenic Avian Influenza A(H5N1) in South America. Travel. Med. Infect. Dis. 2024, 59, 102712. [Google Scholar] [CrossRef]
- Thanawongnuwech, R.; Amonsin, A.; Tantilertcharoen, R.; Damrongwatanapokin, S.; Theamboonlers, A.; Payungporn, S.; Nanthapornphiphat, K.; Ratanamungklanon, S.; Tunak, E.; Songserm, T.; et al. Probable tiger-to-tiger transmission of avian influenza H5N1. Emerg. Infect. Dis. 2005, 11, 699–701. [Google Scholar] [CrossRef]
- Leguia, M.; Garcia-Glaessner, A.; Muñoz-Saavedra, B.; Juarez, D.; Barrera, P.; Calvo-Mac, C.; Jara, J.; Silva, W.; Ploog, K.; Amaro, L.; et al. Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Commun. 2023, 14, 5489. [Google Scholar] [CrossRef] [PubMed]
- Rabalski, L.; Milewska, A.; Pohlmann, A.; Gackowska, K.; Lepionka, T.; Szczepaniak, K.; Swiatalska, A.; Sieminska, I.; Arent, Z.; Beer, M.; et al. Emergence and potential transmission route of avian influenza A (H5N1) virus in domestic cats in Poland, June 2023. Euro Surveill. 2023, 28, 2300390. [Google Scholar] [CrossRef] [PubMed]
- Canadian Food Inspection Agency. Domestic dog tests positive for avian influenza. In Canadian Report. Available online: https://www.canada.ca/en/food-inspection-agency/news/2023/04/domestic-dog-tests-positive-for-avian-influenza-in-canada.html (accessed on 4 April 2023).
- FDA Investigating Raw Pet Food Contaminated with H5N1 Virus; U.S. Food & Drug Administration: Silver Spring, MD, USA, 2024. Available online: https://www.fda.gov/food/alerts-advisories-safety-information/investigation-avian-influenza-h5n1-virus-dairy-cattle (accessed on 3 March 2025).
- CDC. Current Situation: Bird Flu in Dairy Cows & Cats. Centers for Disease Control and Prevention; 2024–2025. Available online: https://www.cdc.gov/bird-flu/risk-factors/bird-flu-in-pets.html (accessed on 3 March 2025).
- Science Alert. Bird Flu Confirmed in Cat Deaths Linked to Raw Pet Food & Milk. 2025. Available online: https://www.sciencealert.com/bird-flu-confirmed-in-cat-deaths-linked-to-raw-pet-food-milk (accessed on 3 March 2025).
- Coleman, K.K.; Bemis, I.G. Avian Influenza Virus Infections in Felines: A Systematic Review of Two Decades of Literature. In Open Forum Infectious Diseases; Oxford University Press: New York, NY, USA, 2025; Volume 12, No. 5. [Google Scholar] [CrossRef]
- Kim, I.H.; Nam, J.H.; Kim, C.K.; Choi, Y.J.; Lee, H.; An, B.M.; Lee, N.J.; Jeong, H.; Lee, S.Y.; Yeo, S.G.; et al. Pathogenicity of Highly Pathogenic Avian Influenza A(H5N1) Viruses Isolated from Cats in Mice and Ferrets, South Korea, 2023. Emerg. Infect. Dis. 2024, 30, 2033–2041. [Google Scholar] [CrossRef]
- Frye, E.A.; Nooruzzaman, M.; Cronk, B.; Laverack, M.; de Oliveira, P.S.B.; Caserta, L.C.; Lejeune, M.; Diel, D.G. Isolation of Highly Pathogenic Avian Influenza A(H5N1) Virus from Cat Urine after Raw Milk Ingestion, United States. Emerg. Infect. Dis. 2025, 31, 1636–1639. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guan, L.; Eisfeld, A.J.; Pattinson, D.; Gu, C.; Biswas, A.; Maemura, T.; Trifkovic, S.; Babujee, L.; Presler, R., Jr.; Dahn, R.; et al. Cow’s Milk Containing Avian Influenza A(H5N1) Virus-Heat Inactivation and Infectivity in Mice. N. Engl. J. Med. 2024, 391, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Moncla, L.; Dudas, G.; VanInsberghe, D.; Sukhova, K.; Lloyd-Smith, J.O.; Worobey, M.; Lowen, A.C.; Nelson, M.I. The global H5N1 influenza panzootic in mammals. Nature 2024, 637, 304–313. [Google Scholar] [CrossRef]
- Davila, K.M.S.; Baker, A.L.; Boggiatto, P.M.; Palmer, M.V.; Putz, E.J.; Olsen, S.C.; Zanella, G.C.; Campos, A.; Buckley, A.; Arruda, B. Transmission of highly pathogenic avian influenza H5N1 to calves fed unpasteurized milk from experimentally infected cows. agriRxiv 2025. agriRxiv:20250056937. [Google Scholar] [CrossRef]
- Perez-Acle, T.; Ravello, C.; Rosemblatt, M. Are we cultivating the perfect storm for a human avian influenza pandemic? Biol. Res. 2024, 57, 96. [Google Scholar] [CrossRef]
- Blondin-Brosseau, M.; Zhang, W.; Gravel, C.; Harlow, J.; Li, X.; Nasheri, N. Comparison of Methods for Extraction of Infectious Influenza Virus from Raw Milk Cheeses. J. Food Prot. 2025, 88, 100529. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Helke, D.; Kuryshko, M.; Abdelwhab, E.M. Survivability of H5N1 avian influenza virus in homemade yogurt, cheese and whey. Emerg. Microbes Infect. 2024, 13, 2420731. [Google Scholar] [CrossRef]
- Suarez, D.L.; Goraichuk, I.V.; Killmaster, L.; Spackman, E.; Clausen, N.J.; Colonius, T.J.; Leonard, C.L.; Metz, M.L. Testing of Retail Cheese, Butter, Ice Cream, and Other Dairy Products for Highly Pathogenic Avian Influenza in the US. J. Food Prot. 2025, 88, 100431. [Google Scholar] [CrossRef]
- Browne, L.; Jackson, J.; Adams, L.; Wilson, A. Updated Risk Assessment: Risk to UK Consumers From Highly Pathogenic Avian Influenza (HPAI) H5N1 B3.13 in US Dairy and Beef Products. FSA Res. Evid. 2025, 10. [Google Scholar] [CrossRef]
- CDC. Influenza (flu): Highly Pathogenic Avian Influenza A(H5N1) Virus in Animals: Interim Recommendations for Prevention, Monitoring, and Public Health Investigations; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2024. Available online: https://www.cdc.gov/flu/avianflu/hpai/hpai-interim-recommendations.html (accessed on 5 June 2025).
- CDC Report, Global Human Cases with Influenza A(H5N1), 1997–2025. Available online: https://www.cdc.gov/bird-flu/php/surveillance/chart-epi-curve-ah5n1.html (accessed on 5 June 2025).
- Mena, A.; von Fricken, M.E.; Anderson, B.D. The Impact of Highly Pathogenic Avian Influenza H5N1 in the United States: A Scoping Review of Past Detections and Present Outbreaks. Viruses 2025, 17, 307. [Google Scholar] [CrossRef]
- Bartlett, M.L.; Palese, P.; Davis, M.F.; Vermund, S.H.; Bréchot, C.; Evans, J.D.; Sauer, L.M.; Osterhaus, A.; Pekosz, A.; Nelson, M.; et al. Enhancing the Response to Avian Influenza in the US and Globally. 2025. [Online]. Available online: www.thelancet.com (accessed on 10 June 2025).
- Kim, H.; Webster, R.G.; Webby, R.J. Influenza Virus: Dealing with a Drifting and Shifting Pathogen. Viral Immunol. 2018, 31, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Liedes, O.; Ekström, N.; Haveri, A.; Solastie, A.; Vara, S.; Rijnink, W.F.; Bestebroer, T.M.; Richard, M.; de Vries, R.D.; Lindh, E.; et al. Inactivated Zoonotic Influenza A(H5N8) Vaccine Induces Robust Antibody Responses Against Recent Highly Pathogenic Avian Influenza Clade 2.3.4.4b A(H5N1) Viruses. medRxiv 2025, 2025-02. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, C.; Zhu, J.; Wang, S.; Ao, X.; He, Y.; Chen, H.; Liao, X.; Kong, D.; Zhou, Y.; et al. Avian influenza mRNA vaccine encoding hemagglutinin provides complete protection against divergent H5N1 viruses in specific-pathogen-free chickens. J. Nanobiotechnol. 2025, 23, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Shi, Y.; Ge, H.; Wang, Y.; Lu, L.; Jiang, S.; Wang, Q. Genomic signatures and host adaptation of H5N1 clade 2.3.4.4b: A call for global surveillance and multi-target antiviral strategies. Curr. Res. Microb. Sci. 2025, 8, 100377. [Google Scholar] [CrossRef]
- Abousenna, M.S.; Shafik, N.G.; Abotaleb, M.M. Evaluation of humoral immune response and milk antibody transfer in calves and lactating cows vaccinated with inactivated H5 avian influenza vaccine. Sci. Rep. 2025, 15, 4637. [Google Scholar] [CrossRef]
- Santos, J.J.; Wang, S.; McBride, R.; Adams, L.; Harvey, R.; Zhao, Y.; Wrobel, A.G.; Gamblin, S.; Skehel, J.; Lewis, N.S.; et al. Bovine H5N1 binds poorly to human-type sialic acid receptors. Nature 2025, 640, E18–E20. [Google Scholar] [CrossRef]
- Eisfeld, A.J.; Biswas, A.; Guan, L.; Gu, C.; Maemura, T.; Trifkovic, S.; Wang, T.; Babujee, L.; Dahn, R.; Halfmann, P.J.; et al. A. J. Eisfeld et al. reply. Nature 2025, 640, E28–E29. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Kiso, M.; Yamada, S.; Someya, K.; Onodera, Y.; Yamaguchi, A.; Matsunaga, S.; Jounai, N.; Yamayoshi, S.; Takeshita, F.; et al. Protective effects of an mRNA vaccine candidate encoding H5HA clade 2.3.4.4b against the newly emerged dairy cattle H5N1 virus. eBioMedicine 2024, 109, 105408. [Google Scholar] [CrossRef] [PubMed]
- USDA-APHIS. Situation Report: H5N1 Detection in U.S. Dairy Cattle; U.S. Department of Agriculture–Animal and Plant Health Inspection Service: 2024. Available online: https://www.aphis.usda.gov/sites/default/files/dairy-cattle-hpai-tech-brief (accessed on 5 June 2025).
- Swayne, D.E.; Sims, L.D.; Brown, I.; Harder, T.; Stegeman, A.; Abolnik, C.; Delgado, M.; Awada, L.; Pavade, G.; Torres, G. Strategic challenges in the global control of high pathogenicity avian influenza. Rev. Sci. Tech. 2024, 89–102. [Google Scholar] [CrossRef]
- Lail, A.J.; Vuyk, W.C.; Machkovech, H.; Minor, N.R.; Hassan, N.R.; Dalvie, R.; Emmen, I.E.; Wolf, S.; Kalweit, A.; Wilson, N.; et al. Amplicon sequencing of pasteurized retail dairy enables genomic surveillance of H5N1 avian influenza virus in United States cattle. PLoS ONE 2025, 20, e0325203. [Google Scholar] [CrossRef] [PubMed]
- Robinson-McCarthy, L.R.; Simmons, H.C.; Graber, A.L.; Marble, C.N.; Graudin, G.W.; McCarthy, K.R. Dairy cattle herds mount a characteristic antibody response to highly pathogenic H5N1 avian influenza viruses. bioRxiv 2025. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- USDA. USDA Actions to Protect Livestock Health from Highly Pathogenic H5N1 Avian Influenza. 2024. Available online: https://www.usda.gov/about-usda/news/press-releases/2024/04/24/usda-actions-protect-livestock-health-highly-pathogenic-h5n1-avian-influenza (accessed on 3 November 2024).





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanta, R.N.; Akther, M.; Prodhan, M.A.; Akter, S.H.; Annandale, H.; Sarker, S.; Abraham, S.; Uddin, J.M. Adaptation and Outbreak of Highly Pathogenic Avian Influenza in Dairy Cattle: An Emerging Threat to Humans, Pets, and Peridomestic Animals. Pathogens 2025, 14, 846. https://doi.org/10.3390/pathogens14090846
Shanta RN, Akther M, Prodhan MA, Akter SH, Annandale H, Sarker S, Abraham S, Uddin JM. Adaptation and Outbreak of Highly Pathogenic Avian Influenza in Dairy Cattle: An Emerging Threat to Humans, Pets, and Peridomestic Animals. Pathogens. 2025; 14(9):846. https://doi.org/10.3390/pathogens14090846
Chicago/Turabian StyleShanta, Rifat Noor, Mahfuza Akther, M. Asaduzzaman Prodhan, Syeda Hasina Akter, Henry Annandale, Subir Sarker, Sam Abraham, and Jasim Muhammad Uddin. 2025. "Adaptation and Outbreak of Highly Pathogenic Avian Influenza in Dairy Cattle: An Emerging Threat to Humans, Pets, and Peridomestic Animals" Pathogens 14, no. 9: 846. https://doi.org/10.3390/pathogens14090846
APA StyleShanta, R. N., Akther, M., Prodhan, M. A., Akter, S. H., Annandale, H., Sarker, S., Abraham, S., & Uddin, J. M. (2025). Adaptation and Outbreak of Highly Pathogenic Avian Influenza in Dairy Cattle: An Emerging Threat to Humans, Pets, and Peridomestic Animals. Pathogens, 14(9), 846. https://doi.org/10.3390/pathogens14090846








