Characterization of a New HIV-1 Second-Generation Circulating Recombinant Form CRF173_63A6 in the Jewish Autonomous Region of Russia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Source
2.2. Ethics
2.3. Sequence Analysis
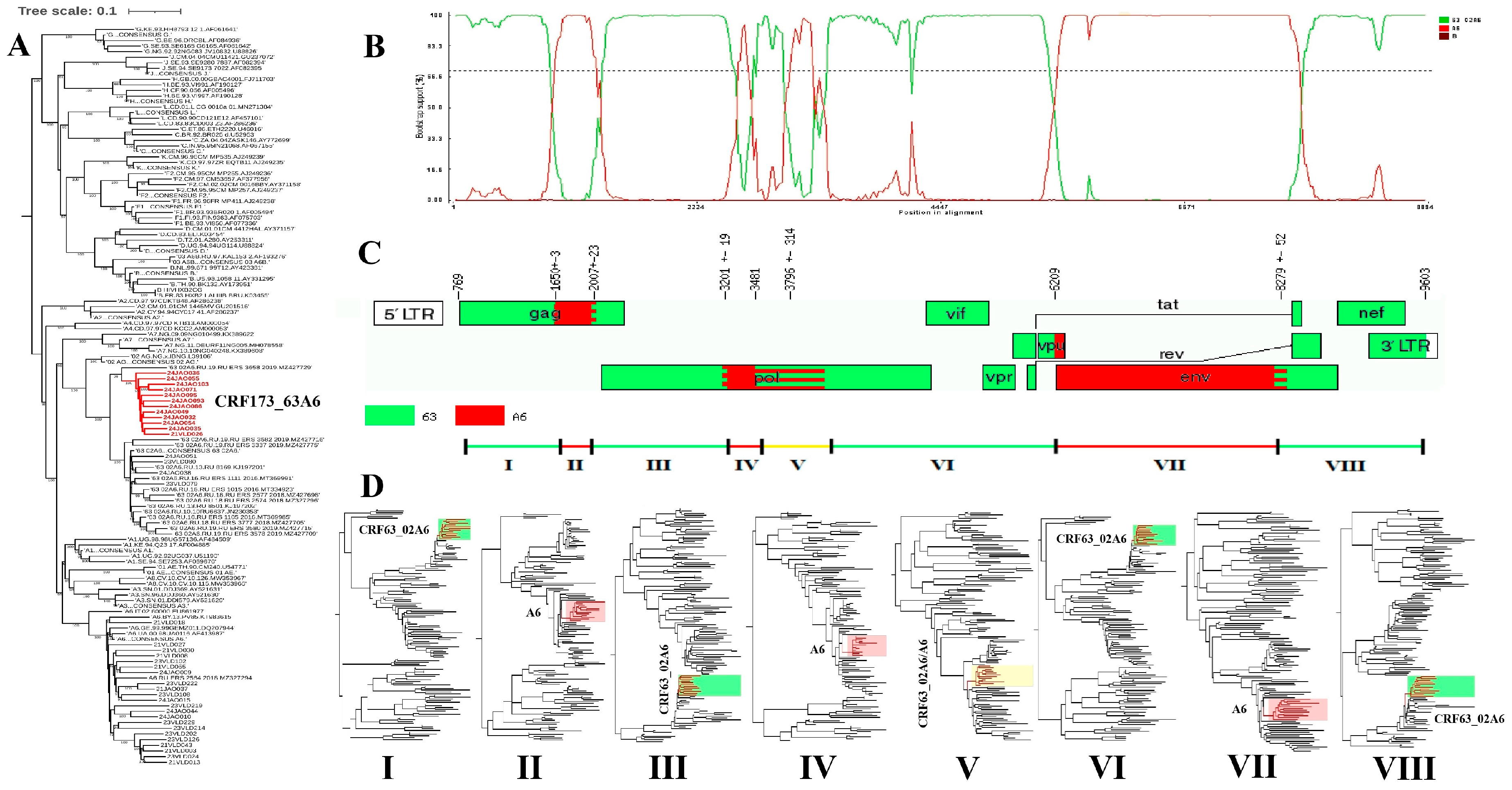
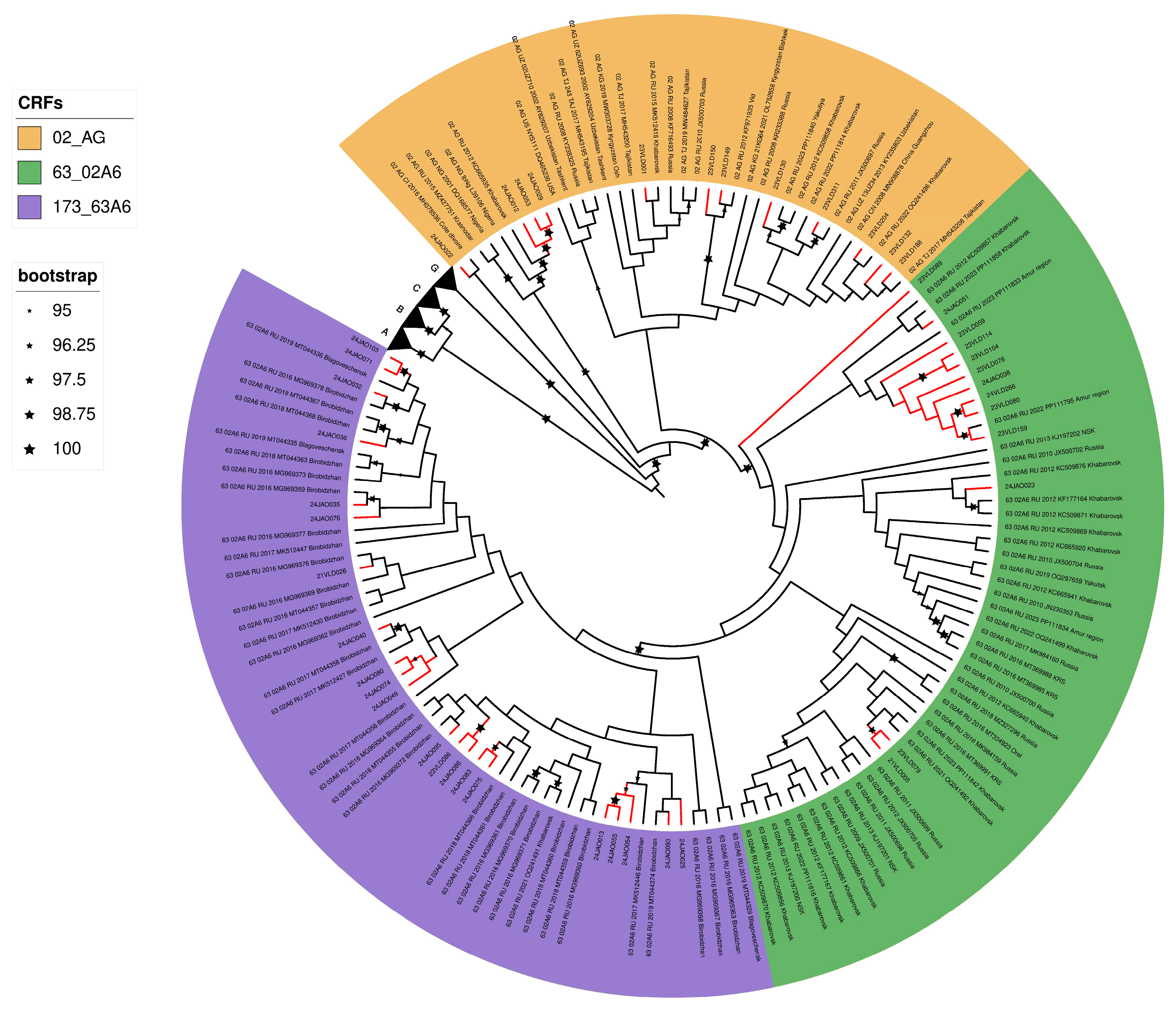
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HIV | Human immunodeficiency viruses |
| JAR | Jewish Autonomous Region |
| AIDS | Acquired Immunodeficiency Syndrome |
| NFLG | Near full-length genome |
| CRF | Circulating recombinant form |
| PWID | People who inject drugs |
| IDU | Injecting drug user |
| URF | Unique recombinant form |
| RIP | Recombinant identification program |
| tMRCA | Time to the most recent common ancestor |
References
- Abidi, S.H.; Aibekova, L.; Davlidova, S.; Amangeldiyeva, A.; Foley, B.; Ali, S. Origin and evolution of HIV-1 subtype A6. PLoS ONE 2021, 16, e0260604. [Google Scholar] [CrossRef]
- Lebedev, A.; Lebedeva, N.; Moskaleychik, F.; Pronin, A.; Kazennova, E.; Bobkova, M. Human immunodeficiency virus-1 diversity in the Moscow region, Russia: Phylodynamics of the most common subtypes. Front. Microbiol. 2019, 10, 320. [Google Scholar] [CrossRef]
- van de Klundert, M.A.A.; Antonova, A.; Di Teodoro, G.; Diez, R.C.; Chkhartishvili, N.; Heger, E.; Kuznetsova, A.; Lebedev, A.; Narayanan, A.; Ozhmegova, E.; et al. Molecular epidemiology of HIV-1 in Eastern Europe and Russia. Viruses 2022, 14, 2099. [Google Scholar] [CrossRef]
- Sivay, M.V.; Maksimenko, L.V.; Osipova, I.P.; Nefedova, A.A.; Gashnikova, M.P.; Zyryanova, D.P.; Ekushov, V.E.; Totmenin, A.V.; Nalimova, T.M.; Ivlev, V.V.; et al. Spatiotemporal dynamics of HIV-1 CRF63_02A6 sub-epidemic. Front. Microbiol. 2022, 13, 946787. [Google Scholar] [CrossRef]
- Safina, K.R.; Sidorina, Y.; Efendieva, N.; Belonosova, E.; Saleeva, D.; Kirichenko, A.; Kireev, D.; Pokrovsky, V.; Bazykin, G.A. Molecular epidemiology of HIV-1 in Oryol oblast, Russia. Virus Evol. 2022, 8, veac044. [Google Scholar] [CrossRef]
- Rudometova, N.B.; Shcherbakova, N.S.; Shcherbakov, D.N.; Mishenova, E.V.; Delgado, E.; Ilyichev, A.A.; Karpenko, L.I.; Thomson, M.M. Genetic Diversity and Drug Resistance Mutations in Reverse Transcriptase and Protease Genes of HIV-1 Isolates from Southwestern Siberia. AIDS Res. Hum. Retroviruses 2021, 37, 716–723. [Google Scholar] [CrossRef]
- Kotova, V.O.; Trotsenko, O.E.; Balakhontseva, L.A.; Bazykina, E.A. Molecular genetic characteristics of HIV-1 variants isolated in the subjects of the Russian far east. Vopr. Virusol. (Probl. Virol. Russ. J.) 2019, 64, 79–89. [Google Scholar] [CrossRef]
- Kirichenko, A.; Kireev, D.; Lapovok, I.; Shlykova, A.; Lopatukhin, A.; Pokrovskaya, A.; Bobkova, M.; Antonova, A.; Kuznetsova, A.; Ozhmegova, E.; et al. HIV-1 drug resistance among treatment-naïve patients in Russia: Analysis of the national database, 2006–2022. Viruses 2023, 15, 991. [Google Scholar] [CrossRef]
- Halikov, M.R.; Ekushov, V.E.; Totmenin, A.V.; Gashnikova, N.M.; Antonets, M.E.; Tregubchak, T.V.; Skliar, L.P.; Solovyova, N.P.; Gorelova, I.S.; Beniova, S.N. Identification of a novel HIV-1 circulating recombinant form CRF157_A6C in Primorsky Territory, Russia. J. Infect. 2024, 88, 180–182. [Google Scholar] [CrossRef]
- Kotova, V.O.; Trotsenko, O.E.; Balakhontseva, L.A.; Bazykina, E.A. The importance of detection of the patterns of HIV-1 genome variability in the system of epidemiological surveillance for HIV-infection (on the example of some territories of the Eastern Federal district). Far East. J. Infect. Pathol. 2019, 37, 48–49. [Google Scholar]
- Kotova, V.O.; Balakhontseva, L.A.; Bazykina, E.A.; Trotsenko, O.E. Circulating recombinant forms of HIV-1 constituent entities of the Far Eastern Federal district. Far East. J. Infect. Pathol. 2021, 40, 79–87. [Google Scholar]
- Koichiro, T.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Kazutaka, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, 78–82. [Google Scholar] [CrossRef]
- Siepel, A.C.; Halpern, A.L.; Macken, C.; Korber, B.T.M. A Computer Program Designed to Screen Rapidly for HIV Type 1 Intersubtype Recombinant Sequences. AIDS Res. Hum. Retroviruses 1995, 11, 1413–1416. [Google Scholar] [CrossRef]
- Schultz, A.-K.; Zhang, M.; Bulla, I.; Leitner, T.; Korber, B.; Morgenstern, B.; Stanke, M. jpHMM: Improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009, 37, W647–W651. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2020, 7, veaa087. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Rhodes, T.; Sarang, A.; Bobrik, A.; Bobkov, E.; Platt, L. HIV transmission and HIV prevention associated with injecting drug use in the Russian Federation. Int. J. Drug Policy 2004, 15, 1–16. [Google Scholar] [CrossRef]
- Rhodes, T.; Lowndes, C.; Judd, A.; Mikhailova, L.A.; Sarang, A.; Rylkov, A.; Tichonov, M.; Lewis, K.; Ulyanova, N.; Alpatova, T.; et al. Explosive spread and high prevalence of HIV infection among injecting drug users in Togliatti City, Russia. Aids 2022, 16, F25–F31. [Google Scholar] [CrossRef]
- Hamers, F.F.; Downs, A.M. HIV in central and eastern Europe. Lancet 2003, 361, 1035–1044. [Google Scholar] [CrossRef]
- Meylakhs, P.; Aasland, A.; Grønningsæter, A. “Until people start dying in droves, no actions will be taken”: Perception and experience of HIV-preventive measures among people who inject drugs in northwestern Russia. Harm Reduct. J. 2017, 14, 33. [Google Scholar] [CrossRef]
- Zyryanova, D.P.; Astakhova, E.M.; Ismailova, T.N.; Bocharov, E.F.; Chernov, A.S.; Totmenin, A.V.; Gashnikova, M.N. Detection of HIV-1 resistant to antiretroviral drugs among tomsk oblast population with newly diagnosed HIV-infection. J. Infectol. 2020, 12, 88–96. [Google Scholar] [CrossRef]
- Maksimenko, L.V.; Sivay, M.V.; Totmenin, A.V.; Shvalov, A.N.; Skudarnov, S.E.; Ostapova, T.S.; Yaschenko, S.V.; Maksutov, R.A.; Gashnikova, N.M. Novel HIV-1 A6/B recombinant forms (CRF133_A6B and URF_A6/B) circulating in Krasnoyarsk region, Russia. J. Infect. 2022, 85, 702–769. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, L.V.; Sivay, M.V.; Antonets, M.E.; Tregubchak, T.V.; Totmenin, A.V.; Skudarnov, S.E.; Ostapova, T.S.; Yashenko, S.V.; Agafonov, A.P.; Gashnikova, N.M. Expanding HIV-1 diversity in Russia: Novel circulating recombinant form between subtypes A6 and B (CRF147_A6B). J. Infect. 2024, 89, 106308. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekushov, V.E.; Halikov, M.R.; Totmenin, A.V.; Antonets, M.E.; Tregubchak, T.V.; Murzin, A.I.; Pavlova, M.N.; Troianova, A.M.; Adusheva, T.P.; Beniova, S.N.; et al. Characterization of a New HIV-1 Second-Generation Circulating Recombinant Form CRF173_63A6 in the Jewish Autonomous Region of Russia. Pathogens 2025, 14, 836. https://doi.org/10.3390/pathogens14090836
Ekushov VE, Halikov MR, Totmenin AV, Antonets ME, Tregubchak TV, Murzin AI, Pavlova MN, Troianova AM, Adusheva TP, Beniova SN, et al. Characterization of a New HIV-1 Second-Generation Circulating Recombinant Form CRF173_63A6 in the Jewish Autonomous Region of Russia. Pathogens. 2025; 14(9):836. https://doi.org/10.3390/pathogens14090836
Chicago/Turabian StyleEkushov, Vasiliy E., Maksim R. Halikov, Alexei V. Totmenin, Mariya E. Antonets, Tatyana V. Tregubchak, Andrey I. Murzin, Marina N. Pavlova, Anastasia M. Troianova, Tatyana P. Adusheva, Svetlana N. Beniova, and et al. 2025. "Characterization of a New HIV-1 Second-Generation Circulating Recombinant Form CRF173_63A6 in the Jewish Autonomous Region of Russia" Pathogens 14, no. 9: 836. https://doi.org/10.3390/pathogens14090836
APA StyleEkushov, V. E., Halikov, M. R., Totmenin, A. V., Antonets, M. E., Tregubchak, T. V., Murzin, A. I., Pavlova, M. N., Troianova, A. M., Adusheva, T. P., Beniova, S. N., Ermolitskaya, A. S., Gorelova, I. S., Agafonov, A. P., & Gashnikova, N. M. (2025). Characterization of a New HIV-1 Second-Generation Circulating Recombinant Form CRF173_63A6 in the Jewish Autonomous Region of Russia. Pathogens, 14(9), 836. https://doi.org/10.3390/pathogens14090836






