Comprehensive Laboratory Analysis of a Scrub Typhus and H1N1 Influenza Co-Infection: A Case Report from Hainan, China
Abstract
1. Introduction
2. Case Presentation
3. Laboratory Studies and Evolutionary Analysis
3.1. Sample Collection and Nucleic Acid Extraction
3.2. Detection and Genetic Analysis of Orientia tsutsugamushi
3.3. Detection and Genetic Analysis of Influenza A Virus
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, G.; Walker, D.H.; Jupiter, D.; Melby, P.C.; Arcari, C.M. A review of the global epidemiology of scrub typhus. PLoS Negl. Trop. Dis. 2017, 11, e0006062. [Google Scholar] [CrossRef] [PubMed]
- Kala, D.; Gupta, S.; Nagraik, R.; Verma, V.; Thakur, A.; Kaushal, A. Diagnosis of scrub typhus: Recent advancements and challenges. 3 Biotech 2020, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.J.; Fuerst, P.A.; Ching, W.M.; Richards, A.L. Scrub typhus: The geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009, 48 (Suppl. S3), S203–S230. [Google Scholar] [CrossRef]
- Banerjee, A.; Kulkarni, S. Orientia tsutsugamushi: The dangerous yet neglected foe from the East. Int. J. Med. Microbiol. 2021, 311, 151467. [Google Scholar] [CrossRef]
- Chunduru, K.; A.R., M.; Poornima, S.; Hande, H.M.; Mridula, M.; Varghese, G.M.; Devaki, R.; Saravu, K. Clinical, laboratory, and molecular epidemiology of Orientia tsutsugamushi infection from Southwestern India. PLoS ONE 2023, 18, e0289126. [Google Scholar] [CrossRef] [PubMed]
- Bonell, A.; Lubell, Y.; Newton, P.N.; Crump, J.A.; Paris, D.H. Estimating the burden of scrub typhus: A systematic review. PLoS Negl. Trop. Dis. 2017, 11, e0005838. [Google Scholar] [CrossRef]
- Win, A.M.; Nguyen, Y.T.H.; Kim, Y.; Ha, N.-Y.; Kang, J.-G.; Kim, H.; San, B.; Kyaw, O.; Htike, W.W.; Choi, D.-O.; et al. Genotypic Heterogeneity of Orientia tsutsugamushi in Scrub Typhus Patients and Thrombocytopenia Syndrome Co-infection, Myanmar. Emerg. Infect. Dis. 2020, 26, 1878–1881. [Google Scholar] [CrossRef]
- Kim, C.-M.; Kim, D.-M.; Yun, N.R. Evaluation of the Diagnostic Accuracy of Antibody Assays for Patients with Scrub Typhus. J. Clin. Microbiol. 2021, 59, e0294220. [Google Scholar] [CrossRef]
- Xin, H.-L.; Yu, J.-X.; Hu, M.-G.; Jiang, F.-C.; Li, X.-J.; Wang, L.-P.; Huang, J.-L.; Wang, J.-F.; Sun, J.-L.; Li, Z.-J. Evaluation of scrub typhus diagnosis in China: Analysis of nationwide surveillance data from 2006 to 2016. Infect. Dis. Poverty 2019, 8, 59. [Google Scholar] [CrossRef]
- Wang, G.; Fu, R.; Zhang, L.; Xue, L.; Al-Mahdi, A.Y.; Xie, X.; Qin, A.; Tang, C.; Du, J.; Huang, Y.; et al. Genomic bacterial load associated with bacterial genotypes and clinical characteristics in patients with scrub typhus in Hainan Island, Southern China. PLoS Negl. Trop. Dis. 2023, 17, e0011243. [Google Scholar] [CrossRef]
- Niu, Y.; Chen, S.; Wang, G.; Guo, Y.; Ge, N.; Hu, X.; Tang, C.; Peng, R.; Cui, X.; Wu, B.; et al. Dual-Genotype Orientia tsutsugamushi Infections, Hainan Island, China, 2023. Emerg. Infect. Dis. 2025, 31, 1222–1225. [Google Scholar] [CrossRef]
- Chang, M.; Shi, S.; Jin, Y.; Wang, G.; Peng, R.; An, J.; Huang, Y.; Hu, X.; Tang, C.; Niu, Y.; et al. Epidemiology and Genetic Evolutionary Analysis of Influenza Virus Among Children in Hainan Island, China, 2021–2023. Pathogens 2025, 14, 142. [Google Scholar] [CrossRef]
- Gupta, N.; Boodman, C.; Jouego, C.G.; Van Den Broucke, S. Doxycycline vs azithromycin in patients with scrub typhus: A systematic review of literature and meta-analysis. BMC Infect. Dis. 2023, 23, 884. [Google Scholar] [CrossRef] [PubMed]
- Varghese, G.M.; Dayanand, D.; Gunasekaran, K.; Kundu, D.; Wyawahare, M.; Sharma, N.; Chaudhry, D.; Mahajan, S.K.; Saravu, K.; Aruldhas, B.W.; et al. Intravenous Doxycycline, Azithromycin, or Both for Severe Scrub Typhus. N. Engl. J. Med. 2023, 388, 792–803. [Google Scholar] [CrossRef]
- Jefferson, T.; Jones, M.A.; Doshi, P.; Del Mar, C.B.; Hama, R.; Thompson, M.J.; Spencer, E.A.; Onakpoya, I.J.; Mahtani, K.R.; Nunan, D.; et al. Neuraminidase inhibitors for preventing and treating influenza in adults and children. Cochrane Database Syst. Rev. 2014, 2018, CD008965. [Google Scholar] [CrossRef]
- Kim, S.W.; Kim, C.M.; Kim, D.M.; Yun, N.R.; Neupane, G.P.; Pyun, S.H.; Yu, B.J. Orientia tsutsugamushi DNA load and genotypes in blood as a marker of severity. Acta Trop. 2021, 215, 105786. [Google Scholar] [CrossRef]
- Coates, B.M.; Staricha, K.L.; Wiese, K.M.; Ridge, K.M. Influenza A Virus Infection, Innate Immunity, and Childhood. JAMA Pediatr. 2015, 169, 956. [Google Scholar] [CrossRef] [PubMed]
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.R.S. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Thiriot, J.D.; Liang, Y.; Gonzales, C.; Sun, J.; Yu, X.; Soong, L. Differential cellular immune responses against Orientia tsutsugamushi Karp and Gilliam strains following acute infection in mice. PLoS Negl. Trop. Dis. 2023, 17, e0011445. [Google Scholar] [CrossRef]
- Jhuria, L.; Muthu, V.; Gupta, S.; Singh, M.P.; Biswal, M.; Goyal, K.; Pannu, A.K.; Kumari, S.; Bhalla, A.; Mohindra, R.; et al. Coinfection of H1N1 Influenza and Scrub Typhus-A Review. QJM 2020, 113, 465–468. [Google Scholar] [CrossRef]
- Yamamoto, C.; Maruyama, A.; Munakata, J.; Matsuyama, T.; Furukawa, K.; Hamashima, R.; Ogawa, M.; Hashimoto, Y.; Fukuda, A.; Inaba, T.; et al. Scrub Typhus and Influenza A Co-Infection: A Case Report. Pathogens 2025, 14, 64. [Google Scholar] [CrossRef]
- Jeong, Y.J.; Kim, S.; Wook, Y.D.; Lee, J.W.; Kim, K.-I.; Lee, S.H. Scrub Typhus: Clinical, Pathologic, and Imaging Findings. Radiographics 2007, 27, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xin, H.; Sun, J.; Lai, S.; Zeng, L.; Zheng, C.; Ray, S.E.; Weaver, N.D.; Wang, L.; Yu, J.; et al. Epidemiologic Changes of Scrub Typhus in China, 1952–2016. Emerg. Infect. Dis. 2020, 26, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xiao, Y.; Wei, X.; Li, X.; Duan, C.; Jia, X.; Jia, R.; Guo, J.; Chen, Y.; Zhang, X.; et al. Spatiotemporal epidemiology and risk factors of scrub typhus in Hainan Province, China, 2011–2020. One Health 2023, 17, 100645. [Google Scholar] [CrossRef]
- Dittrich, S.; Rattanavong, S.; Lee, S.J.; Panyanivong, P.; Craig, S.B.; Tulsiani, S.M.; Blacksell, S.D.; Dance, D.A.; Dubot-Pérès, A.; Sengduangphachanh, A.; et al. Orientia, rickettsia, and leptospira pathogens as causes of CNS infections in Laos: A prospective study. Lancet Glob. Health 2015, 3, e104–e112. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Ou, T.Y.; Chen, F.L.; Hsu, C.W.; Jean, S.S. Co-infection with Orientia tsutsugamushi and Mycoplasma pneumoniae in a traveler. J. Microbiol. Immunol. Infect. 2015, 48, 121–122. [Google Scholar] [CrossRef]
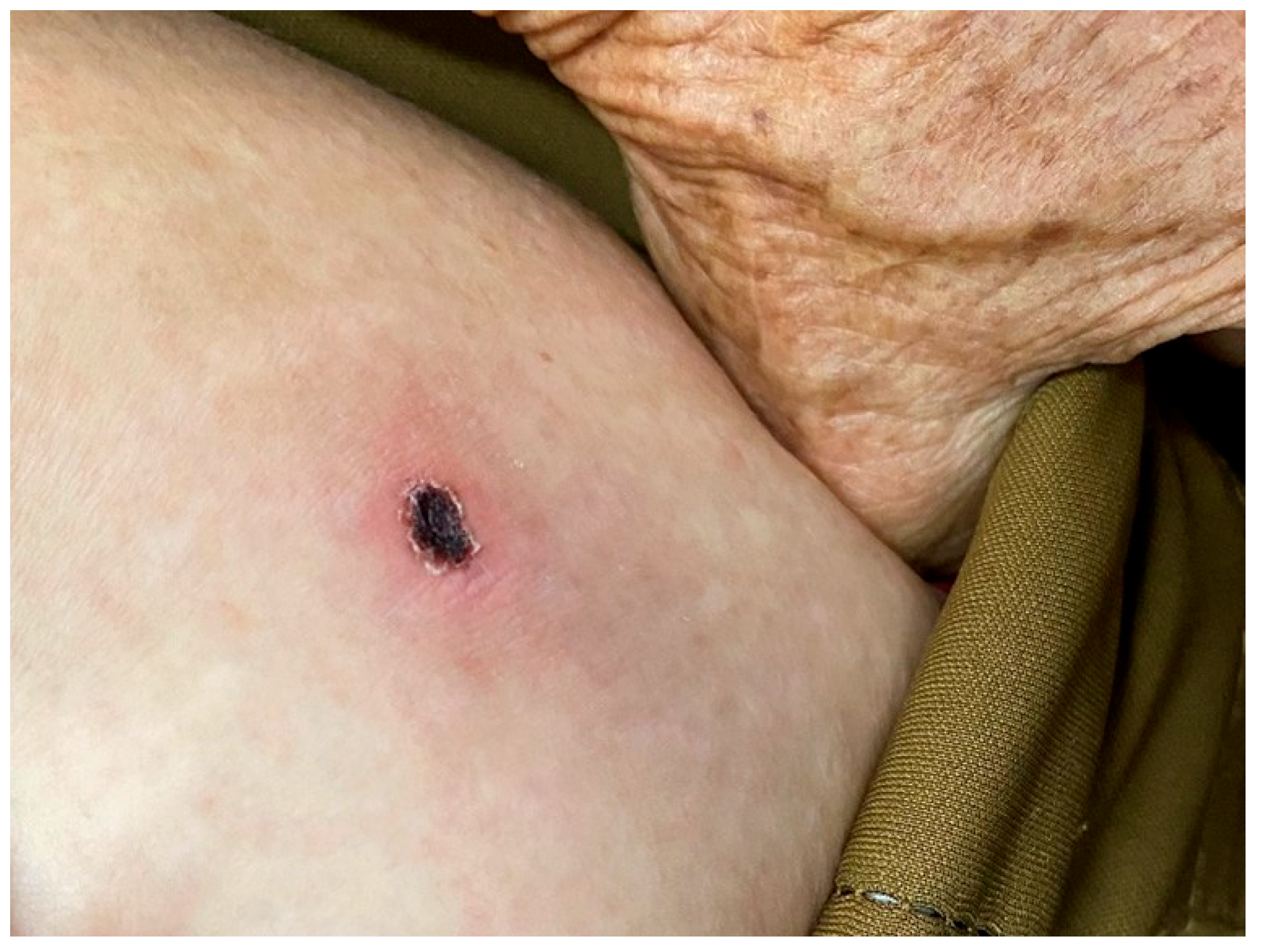
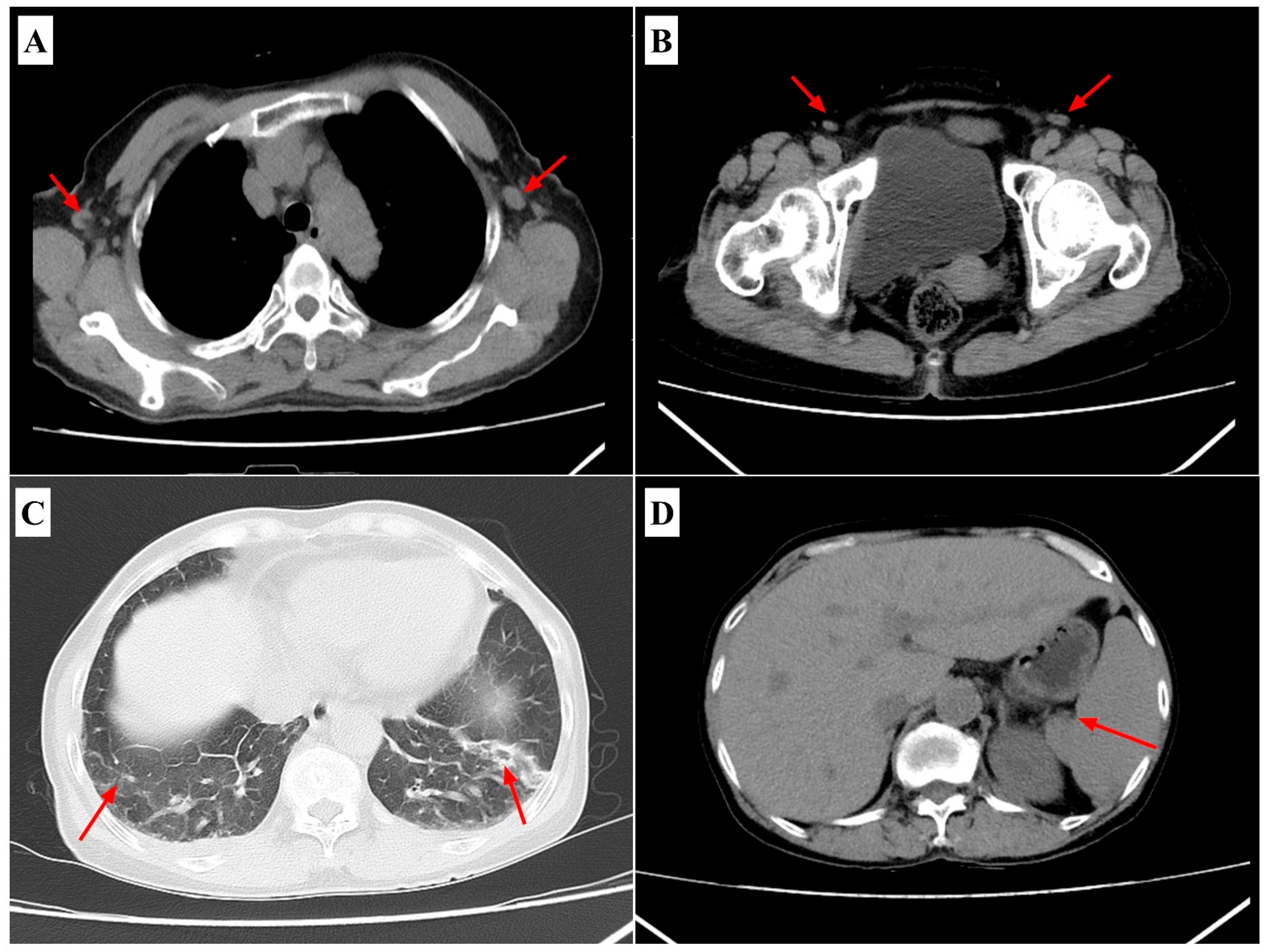
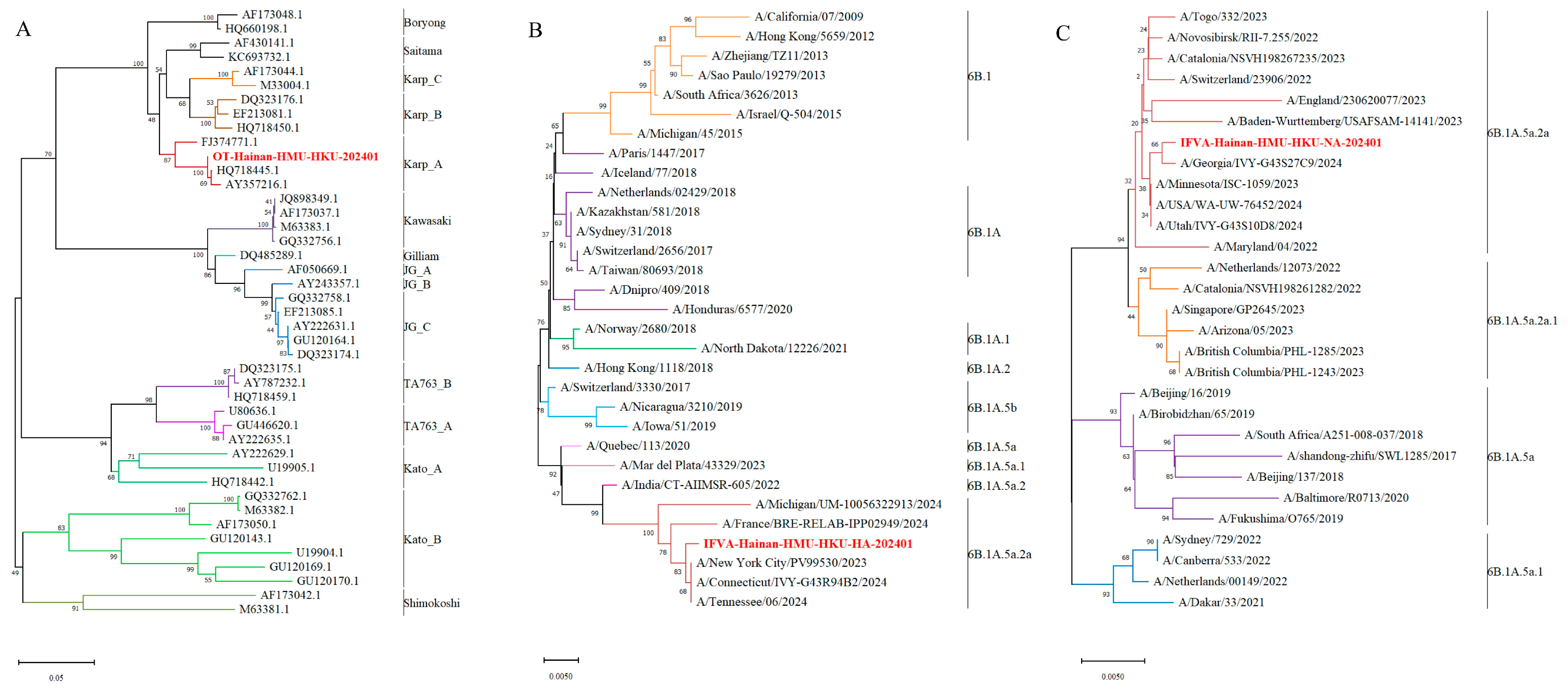
| Category | 22-June Treatment/Peak Phase | 26-June Post-Treatment |
|---|---|---|
| Body temperature (°C) | 39.4 ↑ | 37 |
| hs-CRP (mg/L) | 103.230 ↑ | 9.150 |
| IL-6 (pg/mL) | 121.00 ↑ | Normal |
| PCT (ng/mL) | 1.31 ↑ | 0.18 ↑ |
| Neutrophils Percentage | 76.2% ↑ | 32.0% ↓ |
| Lymphocytes Percentage | 15.4% ↓ | 58.9% ↑ |
| Platelet Count (×103/mL) | 72 ↓ | Normal |
| AST (U/L) | 355 ↑ | 70 ↑ |
| ALT (U/L) | 323 ↑ | 142 ↑ |
| TP (g/L) | 47.5 ↓ | 56.4 ↓ |
| ALB (g/L) | 27.1 ↓ | 30.6 ↓ |
| A/G Ratio | 1.33 | 1.19 ↓ |
| TBIL (μmol/L) | 19.6 ↑ | Normal |
| DBIL (μmol/L) | 12.7 ↑ | Normal |
| APTT (s) | 62.8 ↑ | Normal |
| D-Dimer (μg/mL) | 20.62 ↑ | 1.68 ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Wang, F.; Wu, S.; Chen, Y.; Niu, Y.; Guo, Y.; Lin, D.; Cui, X.; Peng, R.; Xu, Z.; et al. Comprehensive Laboratory Analysis of a Scrub Typhus and H1N1 Influenza Co-Infection: A Case Report from Hainan, China. Pathogens 2025, 14, 810. https://doi.org/10.3390/pathogens14080810
Chen S, Wang F, Wu S, Chen Y, Niu Y, Guo Y, Lin D, Cui X, Peng R, Xu Z, et al. Comprehensive Laboratory Analysis of a Scrub Typhus and H1N1 Influenza Co-Infection: A Case Report from Hainan, China. Pathogens. 2025; 14(8):810. https://doi.org/10.3390/pathogens14080810
Chicago/Turabian StyleChen, Siqi, Fahui Wang, Shannan Wu, Yuanze Chen, Yi Niu, Yijia Guo, Dachuan Lin, Xiuji Cui, Ruoyan Peng, Zhao Xu, and et al. 2025. "Comprehensive Laboratory Analysis of a Scrub Typhus and H1N1 Influenza Co-Infection: A Case Report from Hainan, China" Pathogens 14, no. 8: 810. https://doi.org/10.3390/pathogens14080810
APA StyleChen, S., Wang, F., Wu, S., Chen, Y., Niu, Y., Guo, Y., Lin, D., Cui, X., Peng, R., Xu, Z., Wu, B., Liao, M., Du, Y., Zhang, L., & Yin, F. (2025). Comprehensive Laboratory Analysis of a Scrub Typhus and H1N1 Influenza Co-Infection: A Case Report from Hainan, China. Pathogens, 14(8), 810. https://doi.org/10.3390/pathogens14080810






