Progress in Our Understanding of the Cross-Protection Mechanism of CTV-VT No-SY Isolates Against Homologous SY Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of SY and No-SY Isolates
2.2. Cross-Protection Trials on Sour Orange and Sweet Orange on SO Plants
2.3. Primer Design for RT-PCR HRM Assay
2.4. RNA Isolation and cDNA Synthesis
2.5. Real-Time RT-PCR and HRM Assay
2.6. High-Throughput Amplicon Sequencing of the p23 Gene
3. Results
3.1. Symptomatic Reactions of Sour Orange Seedlings and Sweet Orange Grafted onto SO, Inoculated with No-SY and SY Isolates
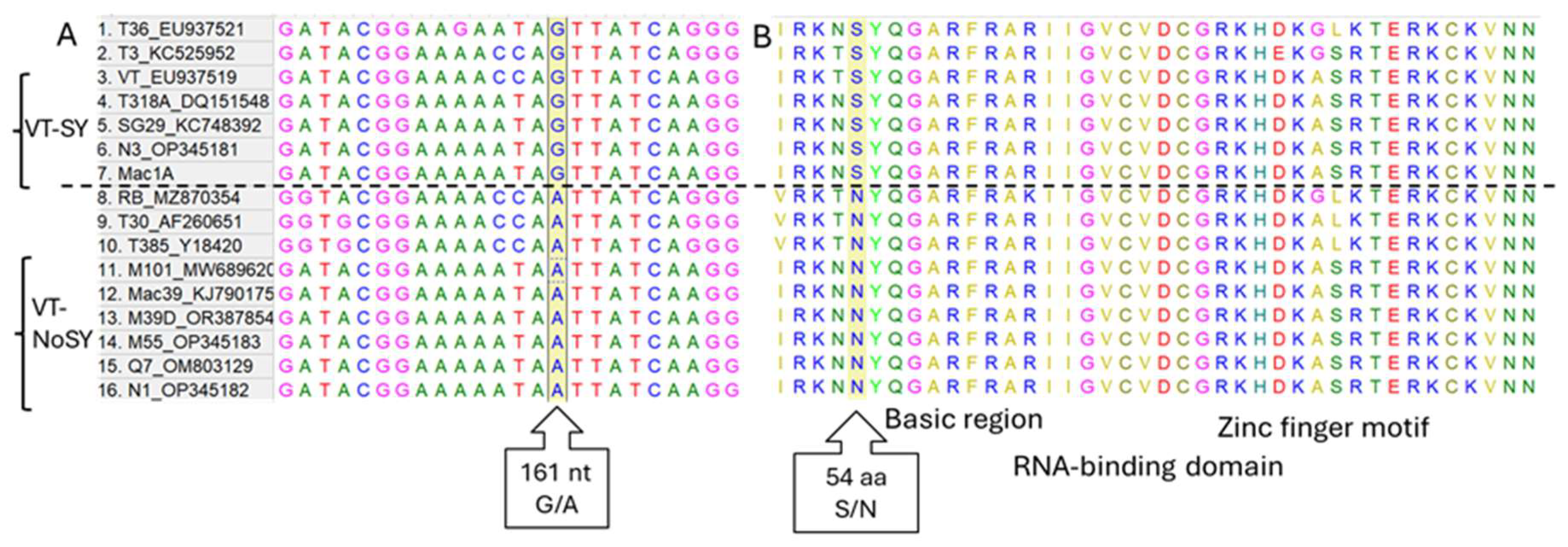
3.2. Analysis of G/A Nucleotide Polymorphism of p23 Gene Among Different CTV Strains
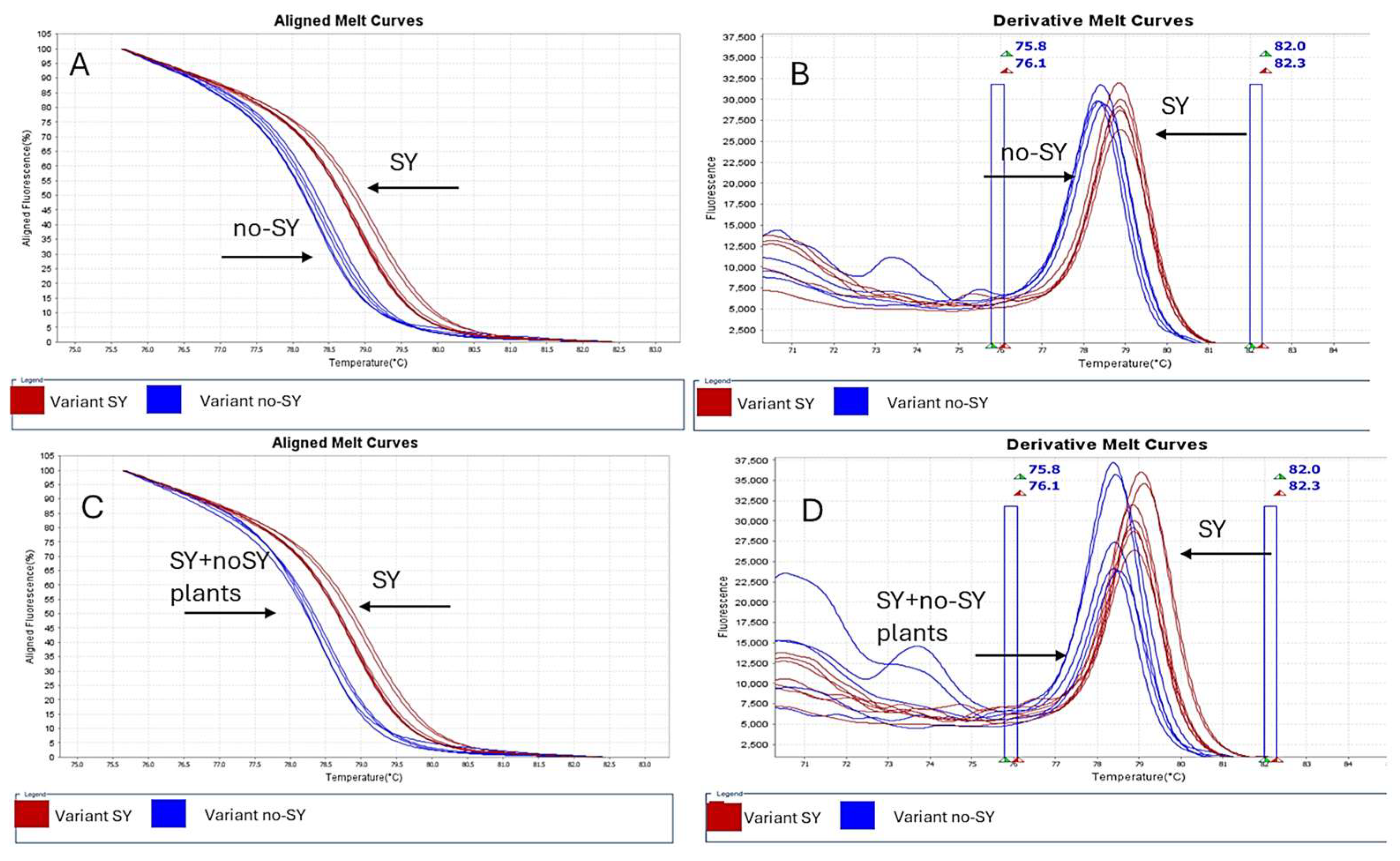
3.3. Evaluation of SY and No-SY VT Variants via Real-Time RT-PCR and HRM Assay on p23 Gene
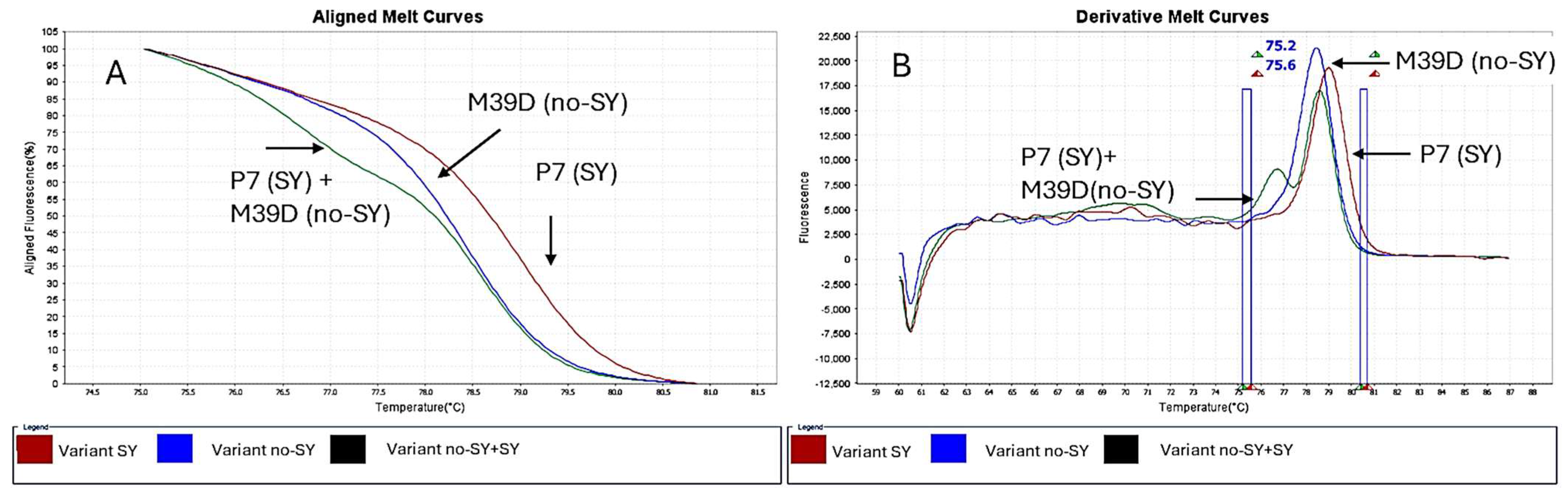
3.4. Real-Time RT-PCR and HRM Analysis of Cross-Protected Plants
3.5. Variant Calling Analysis on p23 Gene of Cross-Protected Sour Orange Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bar-Joseph, M.; Garnsey, S.M.; Gonsalves, D. The closteroviruses: A distinct group of elongated plant viruses. Adv. Virus Res. 1979, 25, 93–168. [Google Scholar] [PubMed]
- Martelli, G.P.; Abou Ghanem-Sabanadzovic, N.; Agranovsky, A.A.; Al Rwahnih, M.; Dolja, V.V.; Dovas, C.; Fuchs, M.; Gugerli, P.; Hu, J.S.; Jelkmann, W.; et al. Taxonomic revision of the family Closteroviridae with special reference to grapevine leafroll associated members of the genus Ampelovirus and the putative species unassigned to the family. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar]
- Moreno, P.; Ambros, S.; Albiach-Marti, M.R.; Guerri, J.; Pena, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef]
- Karasev, A.V.; Boyko, V.P.; Gowda, S.; Nikolaeva, O.V.; Hilf, M.E.; Koonin, E.V.; Niblett, C.L.; Cline, K.; Gumpf, D.J.; Lee, R.F.; et al. Complete sequence of the Citrus tristeza virus RNA genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef]
- Bar-Joseph, M.; Marcus, R.; Lee, R.F. The continuous challenge of Citrus tristeza virus control. Ann. Rev. Phytopathol. 1989, 27, 291–316. [Google Scholar] [CrossRef]
- Marroquín, C.; Olmos, A.; Gorris, M.T.; Bertolini, E.; Martínez, M.C.; Carbonell, E.A.; Hermoso de Mendoza, A.H.; Cambra, M. Estimation of the number of aphids carrying Citrus tristeza virus that visit adult citrus trees. Virus Res. 2004, 100, 101–108. [Google Scholar] [CrossRef]
- Bester, R.; Cook, G.; Maree, H.J. Citrus tristeza virus genotype detection using high-throughput sequencing. Viruses 2021, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Albiach-Martí, M.R.; Robertson, C.J.; Gowda, S.; Tatineni, S.; Belliure, B.; Garnsey, S.M.; Folimonova, S.Y.; Moreno, P.; Dawson, W.O. The pathogenicity determinant of citrus tristeza virus causing the seedling yellows syndrome maps at the 3′-terminal region of the viral genome. Mol. Plant Pathol. 2010, 11, 55–67. [Google Scholar] [CrossRef]
- Bar-Joseph, M.; Catara, A.F.; Licciardello, G. The puzzling phenomenon of seedling yellows recovery and natural spread of asymptomatic infections of citrus tristeza virus: Two sides of the same coin. Hortic. Rev. 2021, 48, 339–362. [Google Scholar] [CrossRef]
- Fagoaga, C.; López, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Viral-like symptoms induced by the ectopic expression of the p23 gene of Citrus tristeza virus are citrus specific and do not correlate with the pathogenicity of the virus strain. Mol. Plant Microbe Interact. 2005, 18, 435–445. [Google Scholar] [CrossRef]
- Ghorbel, R.; López, C.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Transgenic citrus plants expressing the Citrus tristeza virus p23 protein exhibit viral-like symptoms. Mol. Plant Pathol. 2001, 2, 27–36. [Google Scholar] [CrossRef]
- Flores, R.; Moreno, P.; Falk, B.; Martelli, G.P.; Dawson, W.O. e-Book on Closteroviridae. Front. Microbiol. 2013, 4, 411. [Google Scholar] [CrossRef]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Gowda, S.; Ayllón, M.A.; Albiach-Martí, M.R.; Rabindram, R.; Dawson, W.O. The p23 protein of Citrus tristeza virus controls asymmetrical RNA accumulation. J. Virol. 2002, 76, 473–483. [Google Scholar] [CrossRef]
- da Graça, J.V.; van Vuuren, S.P. Managing Citrus tristeza virus losses using cross protection. In Citrus Tristeza Virus Complex and Tristeza Diseases; Karasev, A.V., Hilf, M.E., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2010; pp. 247–260. [Google Scholar]
- Roistacher, C.N.; da Graça, J.V.; Müller, G.W. Cross protection against Citrus tristeza virus—A review. Int. Organ. Citrus Virol. Conf. Proc. (1957–2010) 2010, 17, 1–27. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Robertson, C.J.; Shilts, T.; Folimonov, A.S.; Hilf, M.E.; Garnsey, S.M.; Dawson, W.O. Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J. Virol. 2010, 84, 1314–1325. [Google Scholar] [CrossRef]
- Lee, R.F.; Keremane, M.L. Mild strain cross protection of tristeza: A review of research to protect against decline on sour orange in Florida. Front. Microbiol. 2013, 4, 259. [Google Scholar] [CrossRef]
- Dawson, W.O.; Garnsey, S.M.; Tatineni, S.; Folimonova, S.Y.; Harper, S.J.; Gowda, S. Citrus tristeza virus-host interactions. Front. Microbiol. 2013, 4, 88. [Google Scholar] [CrossRef]
- Folimonova, S.Y. Developing an understanding of cross-protection by citrus tristeza virus. Front. Microbiol. 2013, 4, 76. [Google Scholar] [CrossRef]
- Cook, G.; Coetze, B.; Bester, R.; Breytenbach, J.H.J.; Steyn, C.; de Bruin, R.; Burger, J.T.; Maree, H.J. Citrus tristeza virus isolates of the same genotype differ in stem pitting severity in grapefruit. Plant Dis. 2020, 104, 2362–2368. [Google Scholar] [CrossRef]
- Licciardello, G.; Scuderi, G.; Russo, M.; Bazzano, M.; Bar-Joseph, M.; Catara, A.F. Minor variants of Orf1a, p33, and p23 genes of VT strain citrus tristeza virus isolates show symptomless reactions on sour orange and prevent superinfection of severe VT isolates. Viruses 2023, 15, 2037. [Google Scholar] [CrossRef]
- Fu, S.; Shao, J.; Zhou, C.; Hartung, J.S. Co-infection of sweet orange with severe and mild strains of citrus tristeza virus is overwhelmingly dominated by the severe strain on both the transcriptional and biological levels. Front. Plant Sci. 2017, 8, 1419. [Google Scholar] [CrossRef] [PubMed]
- Sambade, A.; López, C.; Rubio, L.; Flores, R.; Guerri, J.; Moreno, P. Polymorphism of a specific region in gene p23 of citrus tristeza virus allows discrimination between mild and severe isolates. Arch. Virol. 2003, 148, 2325–2340. [Google Scholar] [CrossRef]
- Luttig, M.; van Vuuren, S.P.; van der Vyver, J.B. Differentiation of single aphid cultured sub-isolates of two South African Citrus tristeza virus isolates from grapefruit by single-stranded conformation polymorphism. Int. Organ. Citrus Virol. Conf. Proc. (1957–2010) 2002, 15, 186–196. [Google Scholar]
- Licciardello, G.; Raspagliesi, D.; Bar-Joseph, M.; Catara, A. Characterization of isolates of citrus tristeza virus by sequential analyses of enzyme immunoassays and capillary electrophoresis-single-strand conformation polymorphisms. J. Virol. Methods 2012, 181, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Scuderi, G.; Lombardo, A.; Raspagliesi, D.; Russo, M.; Catara, A.; Licciardello, G. Development and evaluation of a novel probe microarray for genotyping citrus tristeza virus using an integrated Lab-On-Chip device. J. Plant Pathol. 2016, 98, 25–34. [Google Scholar]
- Zablocki, O.; Pietersen, G. Characterization of a novel citrus tristeza virus genotype within three cross-protecting source GFMS12 sub-isolates in South Africa by means of Illumina sequencing. Arch. Virol. 2014, 159, 2133–2139. [Google Scholar] [CrossRef]
- Bester, R.; Cook, G.; Breytenbach, J.H.J.; Steyn, C.; De Bruyn, R.; Maree, H.J. Towards the validation of high-throughput sequencing (HTS) for routine plant virus diagnostics: Measurement of variation linked to HTS detection of citrus viruses and viroids. Virol. J. 2021, 18, 61. [Google Scholar] [CrossRef]
- Licciardello, G.; Scuderi, G.; Ferraro, R.; Giampetruzzi, A.; Russo, M.; Lombardo, A.; Raspagliesi, D.; Bar-Joseph, M.; Catara, A. Deep sequencing and analysis of small RNAs in sweet orange grafted on sour orange infected with two citrus tristeza virus isolates prevalent in Sicily. Arch. Virol. 2015, 160, 2583–2589. [Google Scholar] [CrossRef]
- Pechinger, K.; Chooi, K.M.; MacDiarmid, R.M.; Harper, S.J.; Ziebell, H. A new era for mild strain cross-protection. Viruses 2019, 11, 670. [Google Scholar] [CrossRef]
- Maina, S.; Donovan, N.J.; Plett, K.; Bogema, D.; Rodoni, B.C. High-throughput sequencing for plant virology diagnostics and its potential in plant health certification. Front. Hortic. 2024, 3, 1388028. [Google Scholar] [CrossRef]
- Seminara, S.; Bennici, S.; Scollo, F.; Di Guardo, M.; La Malfa, S.; Gentile, A.; Distefano, G. High Resolution Melting DNA analysis for the traceability of plants and juices of blond and pigmented sweet orange. J. Agric. Food Res. 2025, 21, 101755. [Google Scholar] [CrossRef]
- Scaglione, D.; Ciacciulli, A.; Gattolin, S.; Caruso, M.; Marroni, F.; Casas, G.L.; Jurman, I.; Licciardello, G.; Catara, A.F.; Rossini, L.; et al. Deep resequencing unveils novel SNPs, InDels, and large structural variants for the clonal fingerprinting of sweet orange [Citrus sinensis (L.) Osbeck]. Plant Genome 2025, 18, e20544. [Google Scholar] [CrossRef]
- Li, J.-H.; Yin, Y.-P.; Zheng, H.-P.; Zhong, M.-Y.; Peng, R.-R.; Wang, B.; Chen, X.-S. A high-resolution melting analysis for genotyping urogenital Chlamydia trachomatis. Diagn. Microbiol. Infect Dis. 2010, 68, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of single nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef]
- Bester, R.; Jooste, A.E.C.; Maree, H.J.; Burger, J.T. Real-time RT-PCR high-resolution melting curve analysis and multiplex RT-PCR to detect and differentiate grapevine leafroll-associated virus 3 variant groups I, II, III and VI. Virol. J. 2012, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, G.; Önelge, N.; Yokomi, R.K.; Abou Kubaa, R.; Savino, V.; Saponari, M. Rapid differentiation of citrus Hop stunt viroid variants by real-time RT-PCR and high resolution melting analysis. Mol. Cell. Probes 2013, 27, 221–229. [Google Scholar] [CrossRef]
- Gori, A.; Cerboneschi, M.; Tegli, S. High-resolution melting analysis as a powerful tool to discriminate and genotype Pseudomonas savastanoi pathovars and strains. PLoS ONE 2012, 7, e30199. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Madesis, P.; Zambounis, A.; Tsaftaris, A. High-resolution melting analysis allowed fast and accurate closed-tube genotyping of Fusarium oxysporum formae speciales complex. FEMS Microbiol. Lett. 2012, 334, 16–21. [Google Scholar] [CrossRef]
- Zambounis, A.; Ganopoulos, I.; Chatzidimopoulos, M.; Tsaftaris, A.; Madesis, P. High-resolution melting approaches towards plant fungal molecular diagnostics. Phytoparasitica 2015, 43, 265–272. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 2013; arXiv:1303.3997v2. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lefouili, M.; Nam, K. The evaluation of Bcftools mpileup and GATK Haplotype Caller for variant calling in non-human species. Sci. Rep. 2022, 12, 11331. [Google Scholar] [CrossRef]
- Suastika, G.; Natsuaki, T.; Terui, H.; Kano, T.; Ieki, H.; Okuda, S. Nucleotide sequence of Citrus tristeza virus seedling yellows isolate. J. Gen. Plant Pathol. 2001, 67, 73–77. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Moreno, P.; Guerri, J.; Ambrós, S. Discrimination between mild and severe Citrus tristeza virus isolates with a rapid and highly specific real-time reverse transcription-polymerase chain reaction method using TaqMan LNA probes. Phytopathology 2009, 99, 307–315. [Google Scholar] [CrossRef]
- Sun, Y.D.; Folimonova, S.Y. The p33 protein of Citrus tristeza virus affects viral pathogenicity by modulating a host immune response. New Phytol. 2019, 221, 2039–2053. [Google Scholar] [CrossRef]
- Kang, S.H.; Sun, Y.D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A long non-coding RNA of Citrus tristeza virus: Role in the virus interplay with the host immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Achor, D.; Bar-Joseph, M. Walking together: Cross-protection, genome conservation, and the replication machinery of Citrus tristeza virus. Viruses 2020, 12, 1353. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Sun, Y.D. Citrus tristeza virus: From pathogen to panacea. Annu. Rev. Virol. 2022, 9, 417–435. [Google Scholar] [CrossRef]
- Ziebell, H.; MacDiarmid, R. Prospects for engineering and improvement of cross-protective virus strains. Curr. Opin. Virol. 2017, 26, 8–14. [Google Scholar] [CrossRef]


| Isolate | GenBank | Symptoms on Indicator Host Plants | |||
|---|---|---|---|---|---|
| Sour Orange (‘SY’) | Mexican Lime (’VC’ and ‘SP’) | Duncan Grapefruit (‘SP’) | Sour Orange (‘SY’) | ||
| M101 | MW689620 | no-SY | + | + | + |
| M39D | OR387854 | no-SY | + | + | + |
| M55 | OP345183 | no-SY | + | + | + |
| Q7 | OM803129 | no-SY | + | + | + |
| Nan1 | P345182 | no-SY | + | + | + |
| P7 | KC748392 | SY | +++ | ++ | +++ |
| Nan3 | OP345181 | SY | +++ | ++ | +++ |
| P1R1 | / * | SY | +++ | ++ | +++ |
| VT Isolate | Host Plant | Year of Inoculation | HRM Tm °C | |
|---|---|---|---|---|
| ID | Variant | |||
| M39D | no-SY | Citrus aurantium | 2020 | 78.37 a * |
| Microcitrus papuana | 2021 | 78.42 a | ||
| M. australasica | 78.28 a | |||
| Fortunella obovata | 78.36 a | |||
| Atalantia ceylanica | 78.54 a | |||
| M39D | no-SY | C. paradisi | 2017 | 78.53 a |
| M101 | C. aurantium | 2020 | 78.64 a | |
| M55 | 78.40 a | |||
| P7 | SY | C. aurantium C. sinensis | 2020 | 79.24 b |
| Nan3 | 79.35 b | |||
| P1R1 | C. volkameriana | 2018 | 79.30 b | |
| Infected Source Plants | Melting Temperature (°C) | ||||||
|---|---|---|---|---|---|---|---|
| Trial | Host | VT Isolate | Biological Replicas | Average | |||
| Seq | SO | M39D + P7 | 78.38 | 78.51 | 78.49 | 78.50 | 78.47 a * |
| M39D | 78.41 | 78.38 | 78.40 | 78.45 | 78.39 a | ||
| P7 | 79.33 | 79.27 | 79.10 | 79.24 | 79.23 b | ||
| Nan | SO | Nan1 + P7 | 78.66 | 78.70 | 78.56 | 78.45 | 78.59 a |
| Nan1 | 78.41 | 78.57 | 78.42 | 78.67 | 78.52 a | ||
| H/AA | H/SO | M39D + P7 | 78.43 | 78.35 | 78.32 | 78.55 | 78.41 a |
| M39D | 78.50 | 78.53 | 78.45 | 78.32 | 78.45 a | ||
| CP | SwO/Mac | M101 + P1R1 | 78.42 | 78.47 | 78.42 | 78.47 | 78.45 a |
| Mac | SwO/Mac | M39D + P1R1 | 78.53 | 78.31 | 78.50 | 78.24 | 78.40 a |
| M39D | 78.40 | 78.53 | 78.38 | 78.27 | 78.40 a | ||
| Source Plants | VT Isolate | HRM Tm °C | |
|---|---|---|---|
| Seq2 | M39D (no-SY) + P7 (SY) | Midveins | 78.72 a * |
| Seq8 | M39D (no-SY) + P7 (SY) | 78.64 a | |
| Seq10 | M39D (no-SY) + P7 (SY) | 78.32 a | |
| CP | M101 (no-SY) + P1R1 (SY) | 78.54 a | |
| Seq2R | M39D (no-SY) + P7 (SY) | Roots | 78.34 a |
| Seq7R | M39D (no-SY) | 78.40 a | |
| Seq12R | M39D (no-SY) | 78.42 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Licciardello, G.; Scuderi, G.; Russo, M.; Bazzano, M.; Paradiso, G.; Bar-Joseph, M.; Catara, A.F. Progress in Our Understanding of the Cross-Protection Mechanism of CTV-VT No-SY Isolates Against Homologous SY Isolates. Pathogens 2025, 14, 701. https://doi.org/10.3390/pathogens14070701
Licciardello G, Scuderi G, Russo M, Bazzano M, Paradiso G, Bar-Joseph M, Catara AF. Progress in Our Understanding of the Cross-Protection Mechanism of CTV-VT No-SY Isolates Against Homologous SY Isolates. Pathogens. 2025; 14(7):701. https://doi.org/10.3390/pathogens14070701
Chicago/Turabian StyleLicciardello, Grazia, Giuseppe Scuderi, Marcella Russo, Marina Bazzano, Giuseppe Paradiso, Moshe Bar-Joseph, and Antonino F. Catara. 2025. "Progress in Our Understanding of the Cross-Protection Mechanism of CTV-VT No-SY Isolates Against Homologous SY Isolates" Pathogens 14, no. 7: 701. https://doi.org/10.3390/pathogens14070701
APA StyleLicciardello, G., Scuderi, G., Russo, M., Bazzano, M., Paradiso, G., Bar-Joseph, M., & Catara, A. F. (2025). Progress in Our Understanding of the Cross-Protection Mechanism of CTV-VT No-SY Isolates Against Homologous SY Isolates. Pathogens, 14(7), 701. https://doi.org/10.3390/pathogens14070701






