Insights into the Regulatory Roles of miRNAs in the Salivary Glands of the Soft Ticks Ornithodoros moubata and Ornithodoros erraticus
Abstract
1. Introduction
2. Materials and Methods
2.1. Small RNA Sequences Dataset
2.2. New Bioinformatics Analysis of the Salivary miRNA
2.3. Validation of miRNA Expression in Salivary Glands by Quantitative RT-PCR
2.4. Computational Prediction of miRNA Targets from Salivary Gland Transcriptome Data
2.5. Enrichment Analysis of Biological Processes of mRNA Target Predicted
2.6. Effect of miRNA Knockdown on O. moubata Biology
2.6.1. Dynamics of miRNA Knockdown in O. moubata Females
2.6.2. Antagomir Treatment in O. moubata Females and Assessment of Phenotypic Effects
2.7. Analysis of mRNA Target Genes Following miRNA Knockdown in O. moubata
2.8. Statistical Analysis
3. Results
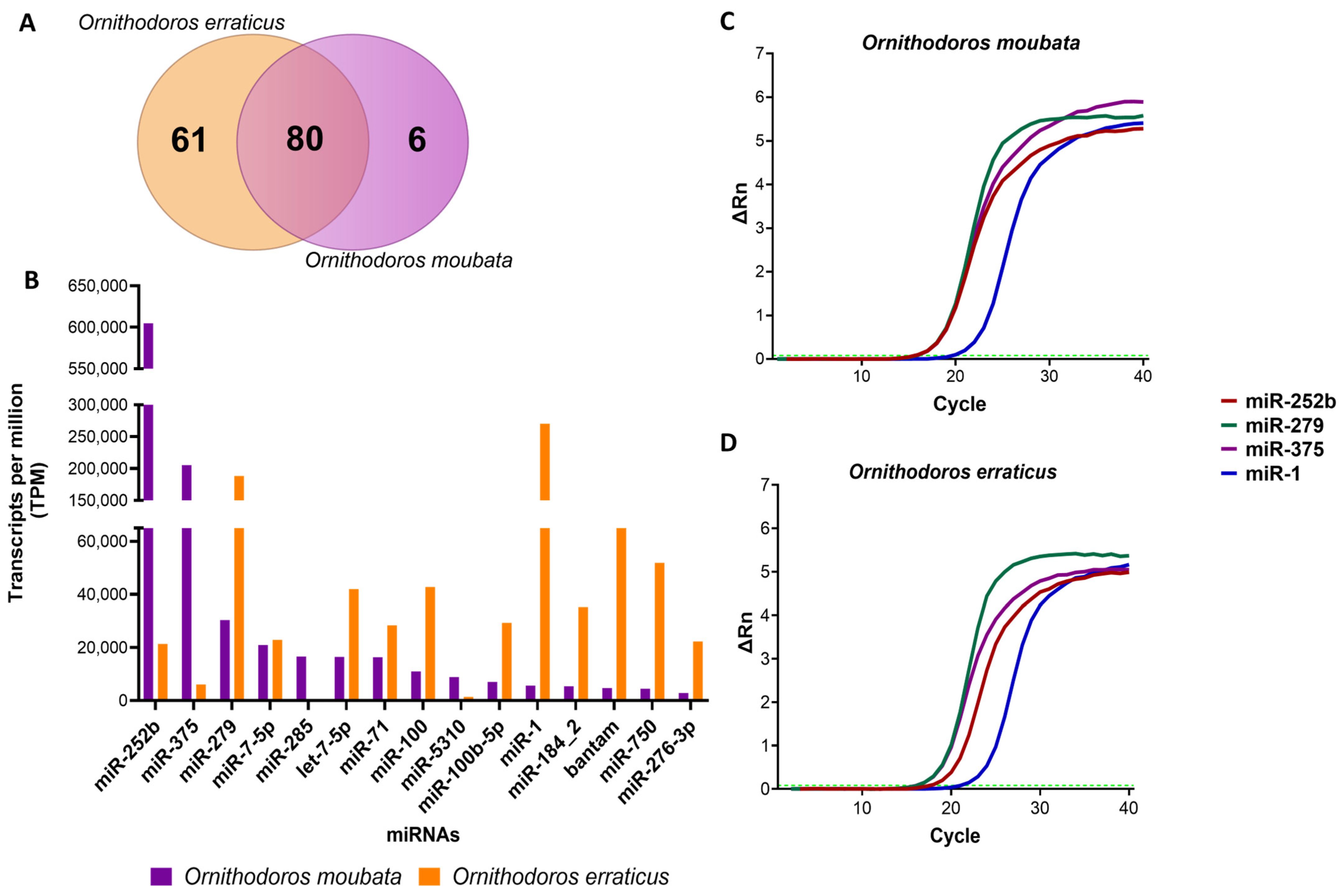
3.1. Analysis of Mature Salivary miRNA Identified in miRBase and Validation of Their Expression in Salivary Gland Tissue
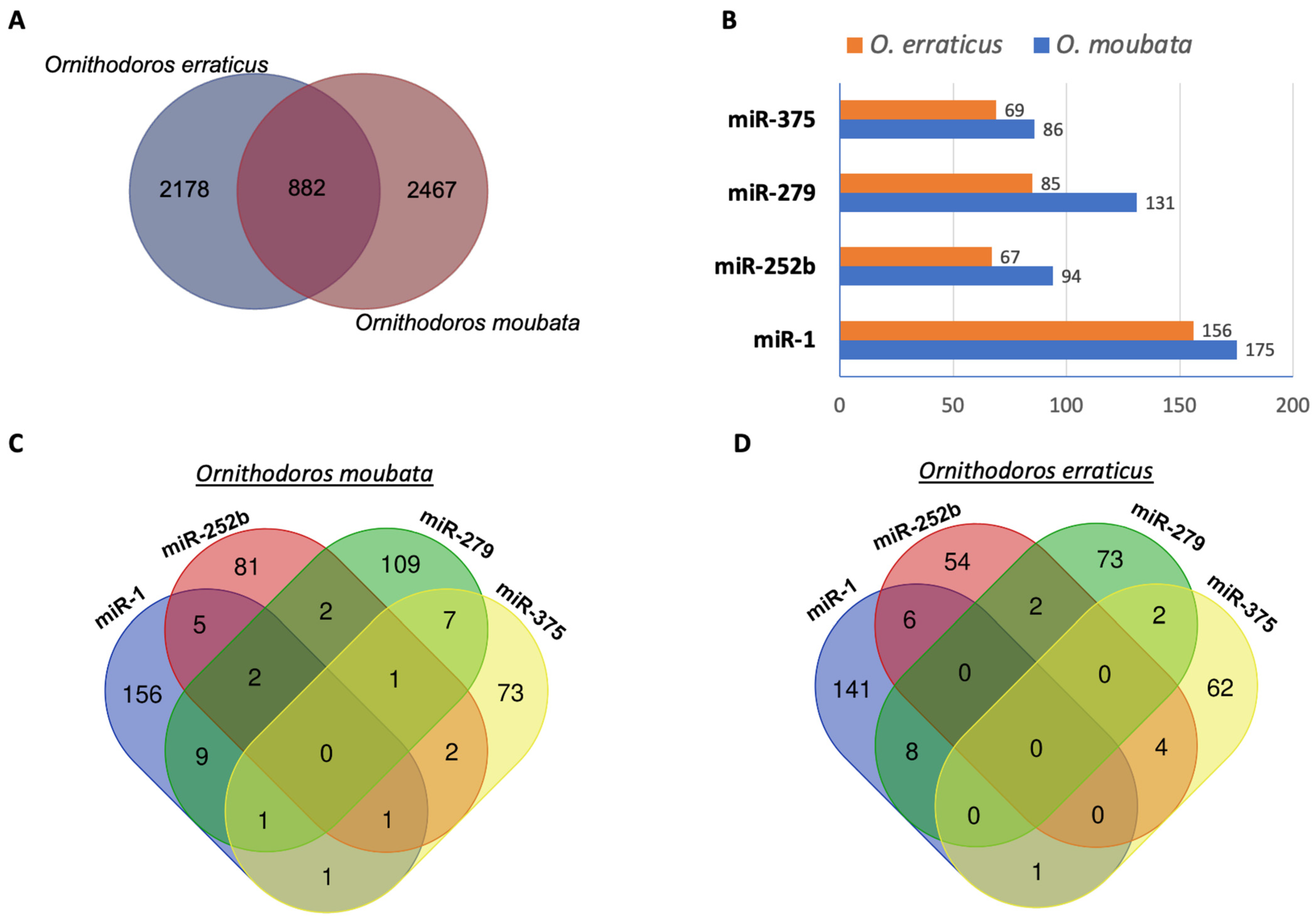
3.2. Prediction of miRNA Target mRNAs from the Salivary Gland Transcriptomes
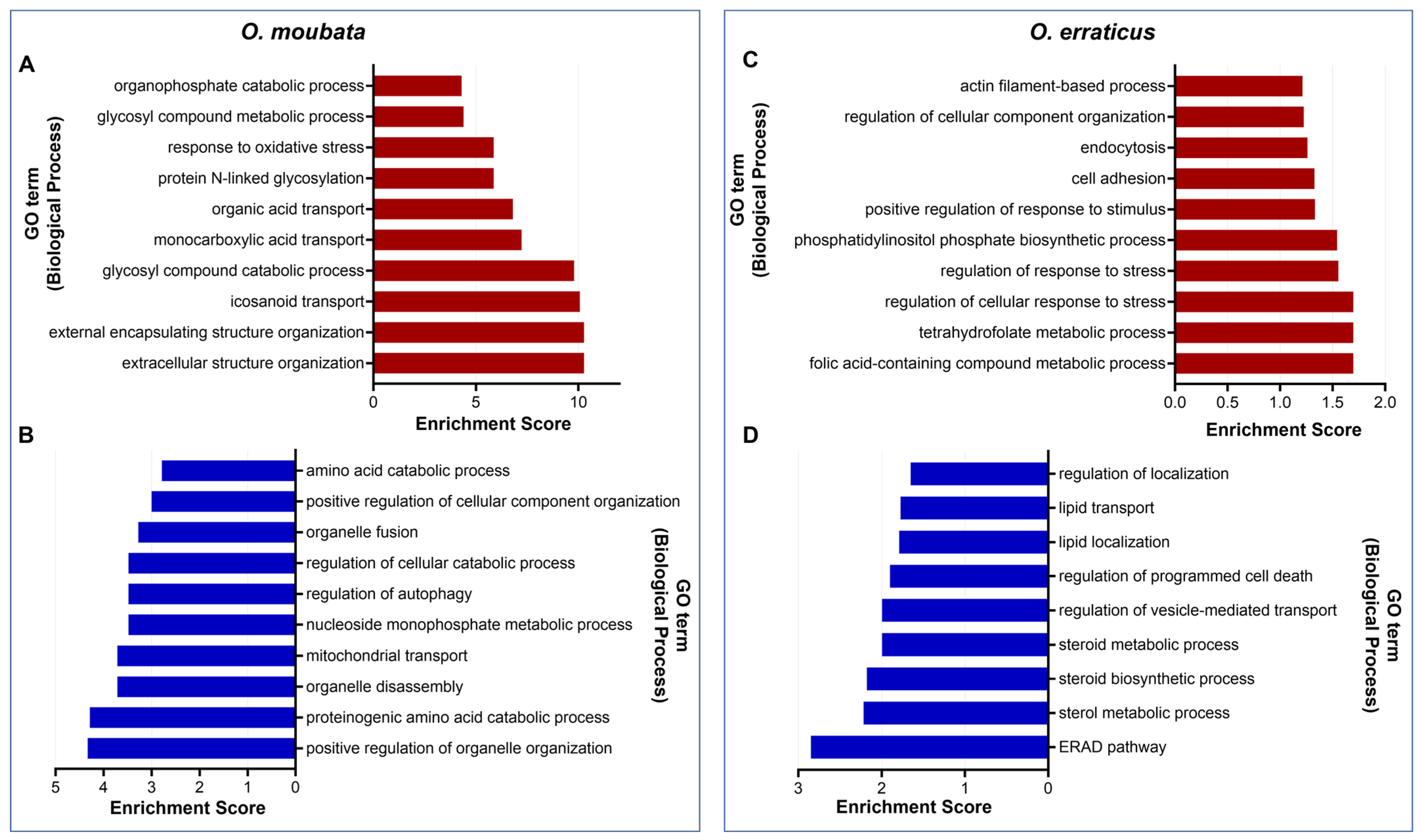
3.3. Functional Analysis of Target Genes
3.4. miRNA Knockdown Effects on the Biological Function of O. moubata
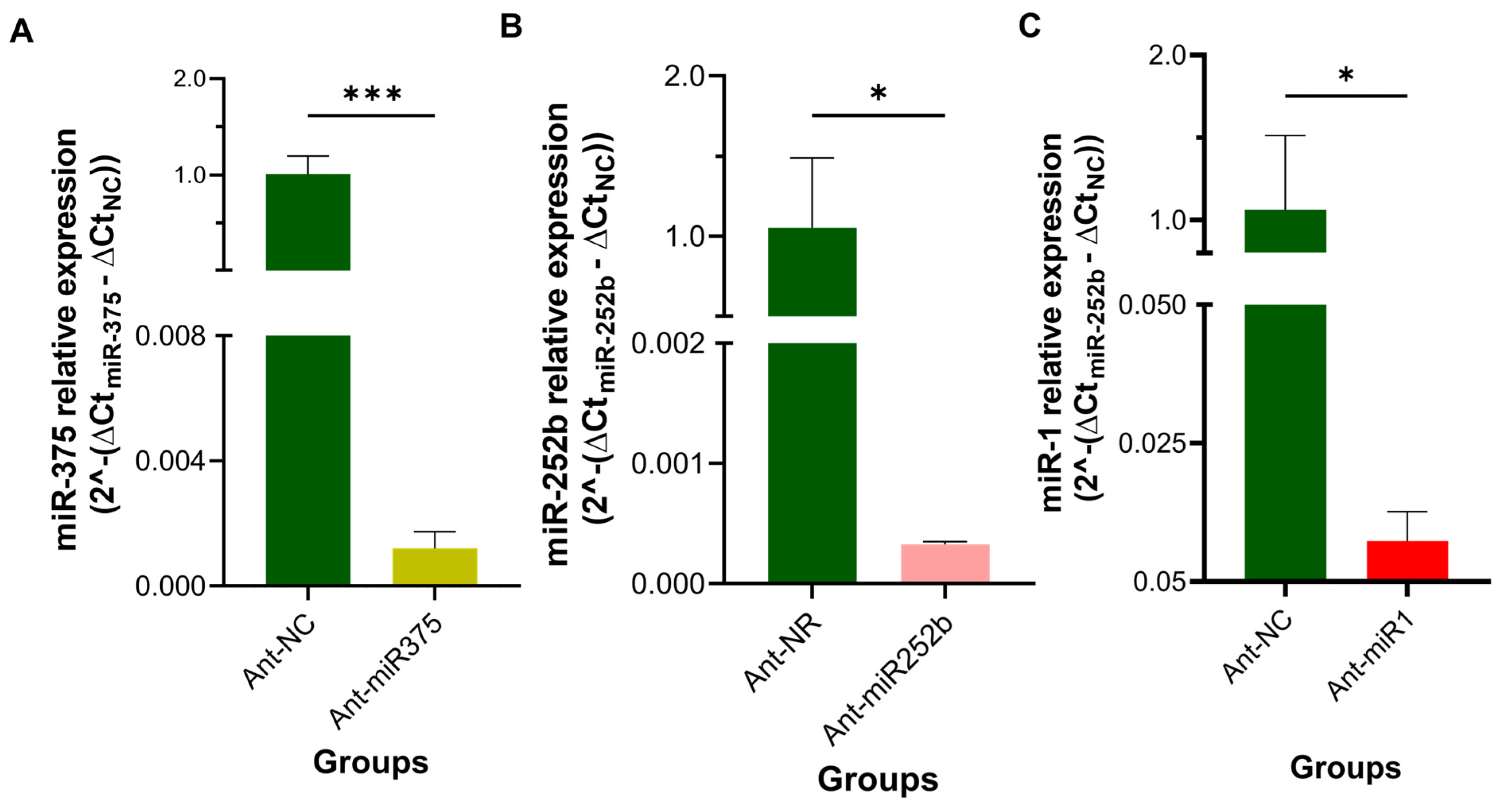
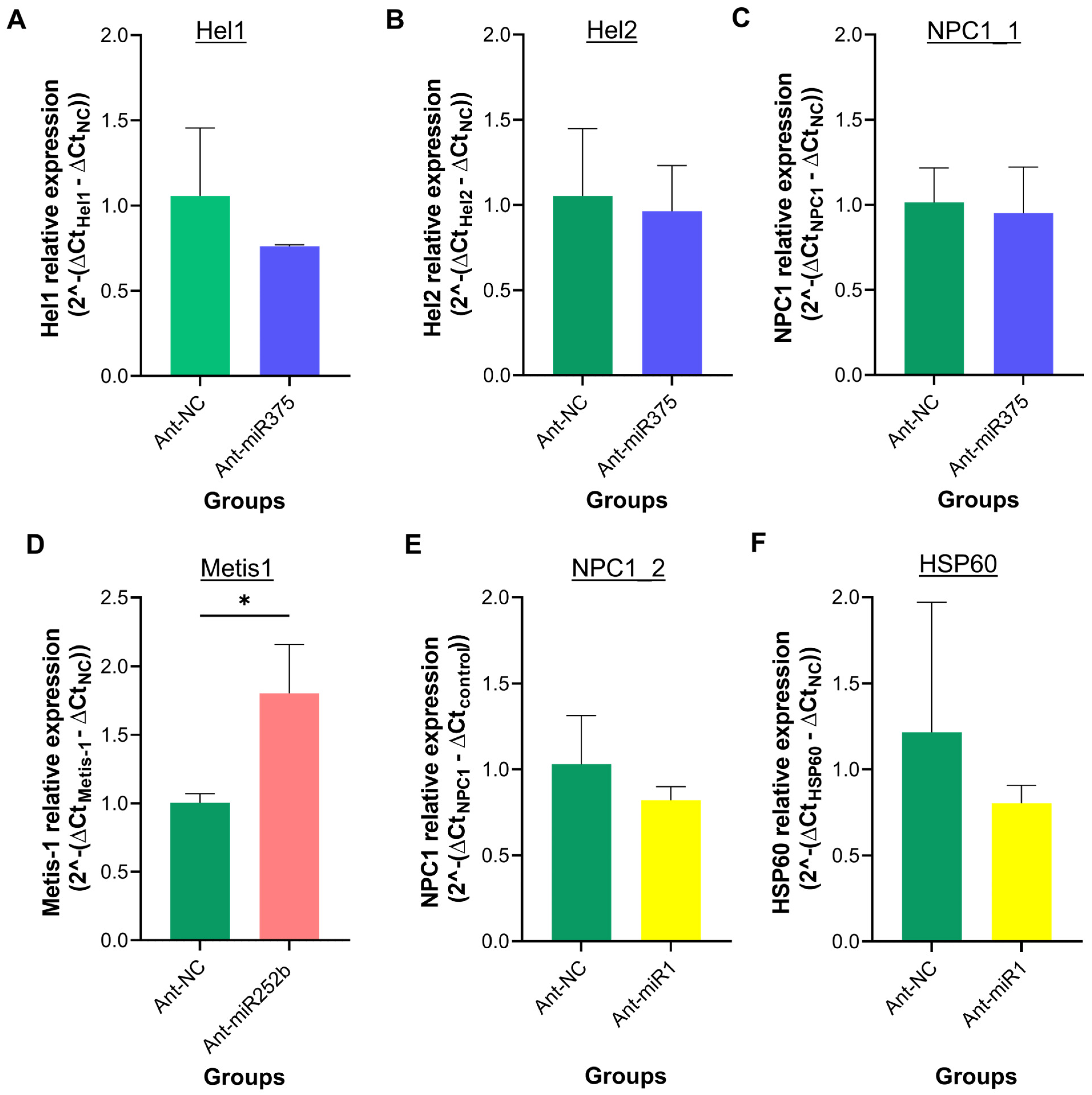
3.5. Quantification of Target Gene Expression Following miRNA Knockdown
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, N. Ticks Biology, Ecology, and Diseases; Academic Press: Cambridge, MA, USA, 2023; 244p. [Google Scholar]
- Ogden, N.H.; Lindsay, L.R. Effects of climate and climate change on vectors and vector-borne diseases: Ticks are different. Trends Parasitol. 2016, 32, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Chung, S.Y. The threat of climate change on tick-borne infections: Rising trend of infections and geographic distribution of climate risk factors associated with ticks. J. Infect. Dis. 2023, 227, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Young, I.; Prematunge, C.; Pussegoda, K.; Corrin, T.; Waddell, L. Tick exposures and alpha-gal syndrome: A systematic review of the evidence. Ticks Tick Borne Dis. 2021, 12, 101674. [Google Scholar] [CrossRef]
- Vaz-Rodrigues, R.; Mazuecos, L.; de la Fuente, J. Current and future strategies for the diagnosis and treatment of the alpha-gal syndrome (AGS). J. Asthma Allergy 2022, 15, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Arcà, B.; Ribeiro, J.M. Saliva of hematophagous insects: A multifaceted toolkit. Curr. Opin. Insect Sci. 2018, 29, 102–109. [Google Scholar] [CrossRef]
- Nuttall, P.A. Wonders of tick saliva. Ticks Tick Borne Dis. 2019, 10, 470–481. [Google Scholar] [CrossRef]
- Nuttall, P.A. Tick saliva and its role in pathogen transmission. Wien. Klin. Wochenschr. 2023, 135, 165–176. [Google Scholar] [CrossRef]
- Aounallah, H.; Bensaoud, C.; M’ghirbi, Y.; Faria, F.; Chmelar, J.I.; Kotsyfakis, M. Tick Salivary Compounds for Targeted Immunomodulatory Therapy. Front. Immunol. 2020, 11, 583845. [Google Scholar] [CrossRef]
- Cano-Argüelles, A.L.; Pérez-Sánchez, R.; Oleaga, A. A microRNA profile of the saliva in the argasid ticks Ornithodoros erraticus and Ornithodoros moubata and prediction of specific target genes. Ticks Tick Borne Dis. 2023, 14, 102249. [Google Scholar] [CrossRef]
- Leal-Galvan, B.; Kumar, D.; Karim, S.; Saelao, P.; Thomas, D.B.; Oliva Chavez, A. A glimpse into the world of microRNAs and their putative roles in hard ticks. Front. Cell Dev. Biol. 2024, 12, 1460705. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Ergin, K.; Çetinkaya, R. Regulation of microRNAs. Methods Mol. Biol. 2022, 2257, 1–32. [Google Scholar]
- Wu, F.; Luo, J.; Chen, Z.; Ren, Q.; Xiao, R.; Liu, W.; Hao, J.; Liu, X.; Guo, J.; Qu, Z.; et al. MicroRNA let-7 regulates the expression of ecdysteroid receptor (ECR) in Hyalomma asiaticum (Acari: Ixodidae) ticks. Parasit. Vectors 2019, 12, 235. [Google Scholar] [CrossRef]
- Liu, W.; Guo, J.; Luo, J.; Ren, Q.; Chen, Z.; Qu, Z.; Wu, Z.; Ni, J.; Xu, X.; Rashid, M.; et al. Analysis of microRNA expression profiles dynamic in different life stages of Haemaphysalis longicornis ticks by deep sequencing of small RNA libraries. Ticks Tick Borne Dis. 2020, 11, 101427. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ren, Q.; Liu, W.; Qiu, X.; Zhang, G.; Tan, Y.; Cao, R.; Yin, H.; Luo, J.; Li, X.; et al. MicroRNA-1 Expression and function in Hyalomma anatolicum anatolicum (Acari: Ixodidae) ticks. Front. Physiol. 2021, 12, 596289. [Google Scholar] [CrossRef]
- Luo, J.; Zhao, S.; Ren, Q.; Wang, Q.; Chen, Z.; Cui, J.; Jing, Y.; Liu, P.; Yan, R.; Song, X.; et al. Dynamic analysis of microRNAs from different life stages of Rhipicephalus microplus (Acari: Ixodidae) by high-throughput sequencing. Pathogens 2022, 11, 1148. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Downs, L.P.; Embers, M.; Flynt, A.S.; Karim, S. Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis. Int. J. Mol. Sci. 2022, 23, 5565. [Google Scholar] [CrossRef]
- Kumar, D.; Budachetri, K.; Rikihisa, Y.; Karim, S. Analysis of Amblyomma americanum microRNAs in response to Ehrlichia chaffeensis infection and their potential role in vectorial capacity. Front. Cell Infect. Microbiol. 2024, 14, 1427562. [Google Scholar] [CrossRef]
- Ramasamy, E.; Taank, V.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Repression of tick microRNA-133 induces organic anion transporting polypeptide expression critical for Anaplasma phagocytophilum survival in the vector and transmission to the vertebrate host. PLoS Genet. 2020, 16, e1008856. [Google Scholar] [CrossRef]
- Hackenberg, M.; Langenberger, D.; Schwarz, A.; Erhart, J.; Kotsyfakis, M. In silico target network analysis of de novo-discovered, tick saliva-specific microRNAs reveals important combinatorial effects in their interference with vertebrate host physiology. RNA 2017, 23, 1259–1269. [Google Scholar] [CrossRef]
- Oleaga-Pérez, A.; Pérez-Sánchez, R.; Encinas-Grandes, A. Distribution and biology of Ornithodoros erraticus in parts of Spain affected by African swine fever. Vet. Rec. 1990, 126, 32–37. [Google Scholar] [PubMed]
- Ribeiro, R.; Otte, J.; Madeira, S.; Hutchings, G.H.; Boinas, F. Experimental infection of Ornithodoros erraticus sensu stricto with two portuguese African swine fever virus strains. Study of factors involved in the dynamics of infection in ticks. PLoS ONE 2015, 10, e0137718. [Google Scholar] [CrossRef] [PubMed]
- Talagrand-Reboul, E.; Boyer, P.H.; Bergström, S.; Vial, L.; Boulanger, N. Relapsing fevers: Neglected tick-borne diseases. Front. Cell Infect. Microbiol. 2018, 8, 98. [Google Scholar] [CrossRef]
- Alotaibi, B.S.; Wu, C.H.; Khan, M.; Nawaz, M.; Chen, C.C.; Ali, A. African swine fever; insights into genomic aspects, reservoirs and transmission patterns of virus. Front. Vet. Sci. 2024, 11, 1413237. [Google Scholar] [CrossRef]
- Oleaga, A.; Soriano, B.; Llorens, C.; Pérez-Sánchez, R. Sialotranscriptomics of the argasid tick Ornithodoros moubata along the trophogonic cycle. PLoS Negl. Trop. Dis. 2021, 15, e0009105. [Google Scholar] [CrossRef]
- Pérez-Sánchez, R.; Carnero-Morán, Á.; Soriano, B.; Llorens, C.; Oleaga, A. RNA-seq analysis and gene expression dynamics in the salivary glands of the argasid tick Ornithodoros erraticus along the trophogonic cycle. Parasit. Vectors 2021, 14, 170. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Huang, Z.; Teeling, E.C. ExUTR: A novel pipeline for large-scale prediction of 3′-UTR sequences from NGS data. BMC Genom. 2017, 18, 847. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Gómez-Martín, C.; Giannoukakos, S.; Medina, J.M.; Scheepbouwer, C.; García-Moreno, A.; Carmona-Saez, P.; Fromm, B.; Pegtel, M.; Keller, A.; et al. sRNAbench and sRNAtoolbox 2022 update: Accurate miRNA and sncRNA profiling for model and non-model organisms. Nucleic Acids Res. 2022, 50, W710–W717. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA Targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Sturm, M.; Hackenberg, M.; Langenberger, D.; Frishman, D. TargetSpy: A supervised machine learning approach for microRNA target prediction. BMC Bioinform. 2010, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Z.; Cao, J.; Li, Y.; Li, H.; Yang, Q.; Zhang, Q.; Liu, X. Identification and evaluation of suitable reference genes for normalization of microRNA expression in Helicoverpa armigera (Lepidoptera: Noctuidae) using quantitative Real-Time PCR. J. Insect Sci. 2017, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Carnero-Morán, Á.; Oleaga, A.; Cano-Argüelles, A.L.; Pérez-Sánchez, R. Function-guided selection of salivary antigens from Ornithodoros erraticus argasid ticks and assessment of their protective efficacy in rabbits. Ticks Tick Borne Dis. 2023, 14, 102218. [Google Scholar] [CrossRef]
- Hussain, M.; Walker, T.; O’Neill, S.L.; Asgari, S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 146–152. [Google Scholar] [CrossRef]
- Šimo, L.; Kazimirova, M.; Richardson, J.; Bonnet, S.I. The essential role of tick salivary glands and saliva in tick feeding and pathogen transmission. Front. Cell Infect. Microbiol. 2017, 7, 275348. [Google Scholar] [CrossRef]
- Neelakanta, G.; Sultana, H. Tick saliva and salivary glands: What do we know so far on their role in arthropod blood feeding and pathogen transmission. Front. Cell Infect. Microbiol. 2022, 11, 816547. [Google Scholar] [CrossRef] [PubMed]
- Bensaoud, C.; Hackenberg, M.; Kotsyfakis, M. Noncoding RNAs in parasite-vector-host interactions. Trends Parasitol. 2019, 35, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, Y.; Cao, J.; Zhang, H.; Yu, Y. Distinctive microRNA profiles in the salivary glands of Haemaphysalis longicornis related to tick blood-feeding. Exp. Appl. Acarol. 2013, 59, 339–349. [Google Scholar] [CrossRef]
- Hao, J.; Luo, J.; Chen, Z.; Ren, Q.; Guo, J.; Liu, X.; Chen, Q.; Wu, F.; Wang, Z.; Luo, J.; et al. MicroRNA-275 and its target Vitellogenin-2 are crucial in ovary development and blood digestion of Haemaphysalis longicornis. Parasit. Vectors 2017, 10, 253. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From mechanism to organism. Front. Cell Dev. Biol. 2020, 8, 537579. [Google Scholar] [CrossRef]
- Barrero, R.A.; Keeble-Gagnère, G.; Zhang, B.; Moolhuijzen, P.; Ikeo, K.; Tateno, Y.; Gojobori, T.; Guerrero, F.D.; Lew-Tabor, A. Evolutionary conserved microRNAs are ubiquitously expressed compared to tick-specific miRNAs in the cattle tick Rhipicephalus (Boophilus) microplus. BMC Genom. 2011, 12, 328. [Google Scholar] [CrossRef]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. miRNA targets: From prediction tools to experimental validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Kern, F.; Krammes, L.; Danz, K.; Diener, C.; Kehl, T.; Küchler, O.; Fehlmann, T.; Kahraman, M.; Rheinheimer, S.; Aparicio-Puerta, E.; et al. Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res. 2021, 49, 127–144. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Lebrón, R.; Rueda, A.; Gómez-Martín, C.; Giannoukakos, S.; Jaspez, D.; Medina, J.M.; Zubkovic, A.; Jurak, I.; Fromm, B.; et al. sRNAbench and sRNAtoolbox 2019: Intuitive fast small RNA profiling and differential expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Barrero, R.A.; Guerrero, F.D.; Black, M.; McCooke, J.; Chapman, B.; Schilkey, F.; Pérez de León, A.A.; Miller, R.J.; Bruns, S.; Dobry, J.; et al. Gene-enriched draft genome of the cattle tick Rhipicephalus microplus: Assembly by the hybrid Pacific Biosciences/Illumina approach enabled analysis of the highly repetitive genome. Int. J. Parasitol. 2017, 47, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Hermance, M.E.; Widen, S.G.; Wood, T.G.; Thangamani, S. Ixodes scapularis salivary gland microRNAs are differentially expressed during Powassan virus transmission. Sci. Rep. 2019, 9, 13110. [Google Scholar] [CrossRef]
- Malik, M.I.; Nawaz, M.; Hassan, I.A.; Zhang, H.; Gong, H.; Cao, J.; Zhou, Y.; Zhou, J. A microRNA profile of saliva and role of miR-375 in Haemaphysalis longicornis (Ixodida: Ixodidae). Parasit. Vectors 2019, 12, 68. [Google Scholar] [CrossRef]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA–target interactions— beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef] [PubMed]
- de Dios-Blázquez, L.; Cano-Argüelles, A.L.; Pérez-Sánchez, R.; González-Sánchez, M.; Oleaga, A. First data on cholesterol metabolism in Ornithodoros argasid ticks: Molecular and functional characterization of the N-terminal domain of Niemann-Pick C1 proteins. Ticks Tick Borne Dis. 2024, 15, 102382. [Google Scholar] [CrossRef] [PubMed]
- Chmelař, J.; Kotál, J.; Kovaříková, A.; Kotsyfakis, M. The use of tick salivary proteins as novel therapeutics. Front Physiol. 2019, 10, 812. [Google Scholar] [CrossRef]
- Decrem, Y.; Mariller, M.; Lahaye, K.; Blasioli, V.; Beaufays, J.; Zouaoui Boudjeltia, K.; Vanhaeverbeek, M.; Cérutti, M.; Brossard, M.; Vanhamme, L.; et al. The impact of gene knock-down and vaccination against salivary metalloproteases on blood feeding and egg laying by Ixodes ricinus. Int. J. Parasitol. 2008, 38, 549–560. [Google Scholar] [CrossRef]

| Ornithodoros moubata | Ornithodoros erraticus | |||
|---|---|---|---|---|
| Read Counts | % of Annotated | Read Counts | % of Annotated | |
| Annotated | 477,439 | - | 1,152,801 | - |
| Ixodes scapularis (42) | 435,567 | 91.2 | 925,853 | 80.3 |
| Rhipicephalus microplus (24) | 2 | 0 | 0 | 0 |
| Parasteatoda tepidariorum (257) | 27,438 | 5.8 | 166,779 | 14.5 |
| Tetranychus urticae (92) | 11,784 | 2.5 | 39,811 | 3.5 |
| Drosophila melanogaster (471) | 1221 | 0.26 | 11,072 | 1.0 |
| Homo sapiens (2693) | 1427 | 0.3 | 9286 | 0.8 |
| Unannotated | 33,542,931 | - | 36,405,630 | - |
| Ornithodoros moubata | Ornithodoros erraticus | |
|---|---|---|
| Number of transcripts * | 25,366 | 18,487 |
| Number of predicted 3’ UTRs | 10,788 | 8487 |
| Target consensus | 11,878 | 13,571 |
| Unique mRNA targets | 3901 | 3633 |
| Unique genes | 3349 | 3060 |
| Parameter | Ant-NC (Control) | Ant-miR252b (% Reduction) | Ant-miR375 (% Reduction) | Ant-miR1 (% Reduction) |
|---|---|---|---|---|
| Ingested blood (mg) | 174.4 ± 7.9 | 174.3 ± 8.0 | 154.2 ± 12.4 (11.5) ** | 151.1 ± 5.2 (13.2) ** |
| Survival (%) | 97.8 ± 3.64 | 91.1 ± 3.3 (6.7) ** | 100 | 100 |
| Oviposition (eggs/female) | 200.4 ± 8.8 | 186.0 ± 12.7 (7.1) | 170.8 ± 15.4 (15.1) ** | 170.4 ± 14.6 (15.2) ** |
| Fertility (nymphs/female) | 190.6 ± 10.6 | 172.3 ± 13.1 (9.9) | 165.5 ± 15.4 (13.4) * | 161.3 ± 15.5 (15.7) * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano-Argüelles, A.L.; Pérez-Sánchez, R.; Gallardo-Escárate, C.; Vizcaíno-Marín, R.; González-Sánchez, M.; Oleaga, A. Insights into the Regulatory Roles of miRNAs in the Salivary Glands of the Soft Ticks Ornithodoros moubata and Ornithodoros erraticus. Pathogens 2025, 14, 595. https://doi.org/10.3390/pathogens14060595
Cano-Argüelles AL, Pérez-Sánchez R, Gallardo-Escárate C, Vizcaíno-Marín R, González-Sánchez M, Oleaga A. Insights into the Regulatory Roles of miRNAs in the Salivary Glands of the Soft Ticks Ornithodoros moubata and Ornithodoros erraticus. Pathogens. 2025; 14(6):595. https://doi.org/10.3390/pathogens14060595
Chicago/Turabian StyleCano-Argüelles, Ana Laura, Ricardo Pérez-Sánchez, Cristian Gallardo-Escárate, Rocío Vizcaíno-Marín, María González-Sánchez, and Ana Oleaga. 2025. "Insights into the Regulatory Roles of miRNAs in the Salivary Glands of the Soft Ticks Ornithodoros moubata and Ornithodoros erraticus" Pathogens 14, no. 6: 595. https://doi.org/10.3390/pathogens14060595
APA StyleCano-Argüelles, A. L., Pérez-Sánchez, R., Gallardo-Escárate, C., Vizcaíno-Marín, R., González-Sánchez, M., & Oleaga, A. (2025). Insights into the Regulatory Roles of miRNAs in the Salivary Glands of the Soft Ticks Ornithodoros moubata and Ornithodoros erraticus. Pathogens, 14(6), 595. https://doi.org/10.3390/pathogens14060595










