Abstract
Proteus mirabilis (P. mirabilis) is an opportunistic pathogen involved in urinary tract infections as well as various nosocomial infections. Emerging resistances to beta-lactams in this species complicates potential treatment since it is intrinsically resistant to colistin. Eleven isolates of carbapenem-non-susceptible P. mirabilis were identified in Sousse Hospital, Tunisia, from January 2018 to December 2022. MICs were determined and isolates were sequenced to determine their resistomes, sequence types, virulence factors, and their clonal relationships. Susceptibility testing showed that all isolates were resistant to carbapenems, aminoglycosides, fluoroquinolones, chloramphenicol, and the trimethoprim/sulfamethoxazole combination. They remained susceptible to the aztreonam/avibactam combination. All isolates produced NDM-1 carbapenemase and ArmA 16S rRNA methylase. In addition, one isolate co-produced the blaVEB-6 gene. All isolates belonged to ST135, and phylogenetic analysis revealed that they were closely related. This study described the first outbreak of NDM-1-producing P. mirabilis in Tunisia.
1. Introduction
Antimicrobial resistance (AMR) is a growing global public health crisis, significantly impacting patient outcomes, healthcare costs, and the effectiveness of current therapeutic strategies [1,2]. The increasing prevalence of AMR has led to a surge in difficult-to-treat infections, particularly in hospitalized patients, where multidrug-resistant (MDR) pathogens contribute to prolonged hospital stays, higher morbidity and mortality rates, and limited treatment options [3,4]. Among these pathogens, Proteus mirabilis, a Gram-negative rod-shaped bacterium, has emerged as a significant cause of nosocomial infections, posing serious therapeutic challenges due to its intrinsic and acquired resistance mechanisms [5].
P. mirabilis is notorious for its ability to swarm across agar surfaces in a characteristic bull’s-eye pattern and its urease activity, which raises urinary pH, facilitating the formation of urinary stones [6]. It is a leading cause of urinary tract infections (UTIs) and catheter-associated urinary tract infections (CAUTIs), where it rapidly colonizes catheter surfaces, leading to encrustation and increased risk of renal damage [7,8,9,10,11]. Beyond UTIs, P. mirabilis is an opportunistic pathogen responsible for a wide range of healthcare-associated infections, including respiratory and wound infections, peritonitis, bacteremia, meningoencephalitis, osteomyelitis, and burn infections [5,12,13,14].
The treatment of P. mirabilis infections is increasingly complicated due to its intrinsic resistance to tigecycline and colistin, as well as reduced susceptibility to imipenem [15]. Additionally, the bacterium frequently acquires resistance mechanisms such as extended-spectrum β-lactamases (ESBLs) and carbapenemases from Ambler class A (KPC), class B (VIM, NDM), and class D (OXA-48-like, OXA-23) [16,17,18,19,20,21,22]. Among these, the emergence of New Delhi metallo-β-lactamase (NDM)-producing P. mirabilis is particularly concerning, as NDM enzymes confer resistance to nearly all β-lactams, including carbapenems, either alone or in combination with β-lactamase inhibitors such as vaborbactam and relebactam [23]. This significantly limits available treatment options and contributes to the global AMR crisis.
Besides resistance, P. mirabilis expresses several virulence factors (VFs) associated with its pathogenicity in humans, including biofilm formation, the production of enzymes and cytotoxins, uroepithelial cell adhesion, motility, and iron acquisition systems. Additionally, this bacterium employs a phosphate transport (Pst) system and diverse iron acquisition mechanisms, including proteobactin (Pbt) and nonribosomal peptide synthetase (NRPS)-derived siderophores such as Yersiniabactin. Moreover, it secretes the hemolysins HpmA and HlyA, which act as potent toxins that disrupt host cell integrity [24,25,26].
Given the clinical importance of carbapenemase-producing P. mirabilis, here we report the first clonal dissemination of NDM-producing P. mirabilis in a university hospital in Tunisia. This study investigates the resistome, and clinical characteristics of P. mirabilis isolates from nosocomial infections, highlighting the urgent need for enhanced surveillance and antimicrobial stewardship to mitigate its spread.
2. Materials and Methods
2.1. Study Design and Clinical Isolates
We conducted a retrospective analysis using the Laboratory Information System to review all clinical isolates of Proteus mirabilis collected from various patient samples and hospital wards at the University Hospital of Sahloul (Sousse, Tunisia) between January 2018 and December 2022.
Strains exhibiting resistance to all tested antibiotics (extensively drug-resistant Proteus mirabilis (XDRPm)), as determined by the Vitek-2 System (bioMérieux, Marcy-l’Étoile, France), were further characterized. The routinely tested antibiotics included cephalexin (CFX), ampicillin (AM), amoxicillin/clavulanic acid (AMC), ticarcillin (TIC), piperacillin (PIP), piperacillin/tazobactam (TZP), cefepime (CF), cefoxitin (FOX), cefotaxime (CTX), ceftazidime (CAZ), imipenem (IMP), ertapenem (ERT), meropenem (MP), aztreonam (ATM), amikacin (AN), gentamycin (GM), tobramycin (TM), ciprofloxacin (CIP), levofloxacine (LVX), chloramphenicol (C), tigecycline (TIG), and trimethoprim-sulfamethoxazole (SXT).
For the selected XDRPm strains, clinical data were retrieved from patient medical records, while information on antibiotic consumption during hospitalization was obtained from pharmacy records.
2.2. Bacterial Reidentification, Susceptibility Testing, MIC Determination and Carbapenemase Detection
Species identification was performed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Microflex; Bruker Daltonics, Bremen, Germany) as previously described [27].
Minimal inhibitory concentrations (MICs) to AMX, AMC, TIC, PIP, TZP, CF, CTX, FOX, CAZ, ceftazidime/avibactam (CAZ-AVI), cefiderocol (CEFID), ceftolozane/tazobactam (FZNTAZ), (IMP), imipenem-relebactam (IMP-RL), MP, meropenem/vaborbactam (MP-VAB), ERT, ATM, aztreonam/avibactam (ATM-VAB), AN, GM, TM, CIP, LVX, TEM, C, TIG, SXT, and colistin (COL), were determined using broth microdilution using precoated plates (Thermofisher, Les Ulis, France). MICs were interpreted according to EUCAST breakpoints as updated in 2023 [28]. Carbapenemase detection was performed using the Carba NP test as previously described [29]. The five most prevalent carbapenemase families in Enterobacterales (KPC, NDM, VIM, IMP and OXA-48-like) were searched by means of immunochromatographic assay using the NG-test Carba5 test (NG Biotech, Guipry, France) according to manufacturer’s instructions [30,31].
2.3. Plasmid Identification and Transfer of β-Lactam Resistance Determinants
Plasmid DNA was extracted using the Kieser method as described previously [32]. Plasmids of ca. 154, 66, 48, and 7 kb of Escherichia coli NCTC 50192 were used as plasmid size markers. Plasmid DNA of the isolates was analyzed by means of electrophoresis on a 0.7% agarose gel and we attempted to introduce it via electroporation into E. coli TOP10.
2.4. Whole-Genome Sequencing and Bioinformatics Analysis
Whole-genome sequencing was performed using Illumina’s Nextseq 500 on the PIBNET sequencing platform (Institut Pasteur, Paris, France). Genome assembly was performed using CLC Genomics Workbench v12 (Qiagen, Les Ulis, France). Initial genome annotation was performed using Rapid Annotation using Subsystem Technology (RAST) (https://rast.nmpdr.org/, accessed on 17 October 2023), Center for Genomic Epidemiology (CGE) services (https://www.genomicepidemiology.org/services/, accessed on 17 November 2023) [33]. The sequence type (ST) of isolates was determined according to the MLST schemes available at PubMLST (https://pubmlst.org/, accessed on 13 November 2023). Virulence gene sequences and functions were searched using virulence factors of the pathogenic bacteria database available at (http://www.mgc.ac.cn/VFs/, accessed on 15 November 2023) [28].
To assess population bias, we performed a core-genome-based phylogenetic analysis of our collection, comparing it to unrelated carbapenemase-producing ST135 P. mirabilis isolates from the French National Reference Center. Genome annotation was performed with Prokka v1.14.6, and pan- and core-genome identification was performed with Panaroo v1.5.1 (70% protein identity for pangenome families, 80% presence for core genes). Core genome alignment was performed using MAFFT v7.490. The best-fit model, selected via the Bayesian information criterion (BIC) in ModelFinder Plus, guided phylogenetic inference with IQ-TREE v2.0.7.
3. Results
3.1. Demographic, Clinical, and Microbiological Data
In total, 417 P. mirabilis isolates were recovered in Sahloul University Hospital in Sousse, Tunisia, over a 4-year period (January 2018 to December 2022), of which 262 were ampicillin-resistant (62.2%), 214 were resistant to the amoxicclin/clavulanate combination (51.3%), and 11 were resistant to all antibiotics available for clinical use in our institution (XDRPm). These 11 P. mirabilis isolates displayed a positive CarbaNP, indicating the production of a carbapenemase. The lateral flow immunochromatographic assay NG-test Carba 5 (NG Biotech, Guipry, France) identified the production of an NDM-type carbapenemase.
These 11 P. mirabilis isolates were isolated from patients with several comorbidities who had experienced prolonged hospitalization in the intensive care unit (ICU), with a positive culture being yielded from various clinical specimens. Most of these patients (n = 10/11) were previously exposed to mechanical ventilation and intravascular devices and 6/11 had undergone surgery. Ten patients were treated with broad-spectrum antibiotics including colistin. The demographic and clinical characteristics of these patients and data concerning previous antimicrobial therapy are summarized in Table 1.

Table 1.
Extensively drug-resistant Proteus mirabilis (XDRPm) isolates, including their antibiotic resistance profiles, associated invasive procedures, and patient outcomes.
3.2. Antimicrobial Susceptibility Testings
All isolates were resistant to penicillins, broad-spectrum cephalosporins such as CTX (MIC > 16 mg/L), CAZ (MIC > 16 mg/L), and FEP (MIC > 16 mg/L). Carbapenem resistance was observed, with resistance to IMP and intermediate resistance to MP (MIC of IMP at 32 mg/L and MP at 4 mg/L), except for ERT (MIC ≤ 0.5 mg/L). These isolates were also resistant to the CAZ-AVI combination (MIC > 16 mg/L). All isolates except for P2-109-B1 remained susceptible to ATM and all isolates were susceptible to the ATM-AVI combination. All isolates were resistant to AN and GM (MIC > 32 mg/L), to fluoroquinolones (MIC > 2 mg/L), SXT combinations, C, tcyclines, and COL (intrinsic resistance in P. mirabilis).
3.3. Resistome and Genetic Relatedness
The draft genome of the 11 P. mirabilis isolates revealed an average contig number of 71.9 contigs with an L50 of 8 and an N50 of 179,556 bp. The overall GC content was of 39.1%. The average size of the genome was 4.23 Mb.
All the Illumina-sequenced P. mirabilis isolates carried the blaNDM-1 carbapenemase gene. One isolate (P2-109-B1) also carried the blaVEB-6 ESBL-encoding gene (Table 2). In addition, a wide variety of antibiotic resistance genes against aminoglycosides (aph(6)-Id, aac(6′)-Ib, ant(3″)-Ia, aph(3′)-Ia, armA, aadA2), fluoroquinolones (qnrA1, parC (S84I), parE (K84E), gyrA (S83I), gyrB (E466D)), sulfonamides (sul2, sul1), trimethoprim (dfrA32, dfrA1), macrolides (ere(A), msr(E)), and chloramphenicol (cat, floR) (Table 2) were also observed.

Table 2.
Genomic features and epidemiological insights into multidrug-resistant Proteus mirabilis isolates reported in the present study: analysis of the resistome, virulome, disinfectant resistance, and sequence typing.
A global phylogenetic analysis with P. mirabilis genomes from GenBank revealed that the eleven P. mirabilis isolates were clustered, indicating a potential outbreak (Figure 1A). Accordingly, MLST analysis indicated that all of these isolates belonged to the same sequence type, ST135. To definitively decipher their clonal relationships, a second phylogenetic analysis was performed using epidemiologically unrelated carbapenemase-producing ST135 P. mirabilis isolates recovered from the French National Reference Center (Figure 1B). This analysis demonstrated that these eleven isolates were closely related, indicating potential cross transmission within the hospital.
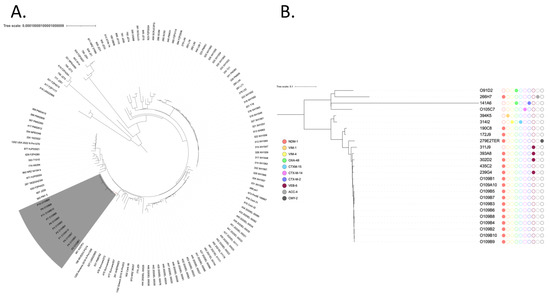
Figure 1.
(A) Phylogenetic analysis of NDM-1-producing P. mirabilis. The eleven isolates recovered from Tunisia are colored in gray. The tree scale is indicated by the horizontal axis. (B) Phylogenetic tree of ST-135 P. mirabilis from Tunisia and from the French National Reference Center for antibiotic resistance: carbapenemase-producing Enterobacterales, Le Kremlin-Bicêtre, France. Broad-spectrum beta-lactamases are indicated by colored circles.
This assumption is further supported by epidemiological data: all cases originated from the same ICU unit, where healthcare personnel and medical staff were shared among patients, increasing the risk of dissemination. Additionally, one affected patient, initially in the neurosurgery department, was later transferred to the POG-ICU, where the other cases were identified. These combined findings suggest a likely intra-hospital spread of the unique clone, although further epidemiological investigations would be necessary to definitively confirm direct transmission events.
3.4. Transfer of Carbapenem Resistance
Kieser-extracted plasmid DNA followed by transformation in E. coli did not allow the transfer of carbapenem resistance, likely indicating that the carbapenemase genes were carried on the chromosome. However, Kieser-extracted plasmid DNA analyzed on a 0.7% agarose gel revealed the presence of an ca. 66 kb plasmid in 9/11 isolates. No plasmid was identified in the strain harboring the blaVEB-6 ESBL gene.
4. Discussion
P. mirabilis is naturally susceptible to all beta-lactams along with aminoglycosides and fluoroquinolones [16]. However, acquired resistance to β-lactams in addition to several classes of antibiotics including SXT, fluroquinolones, fosfomycin, aminoglycosides, and sulfonamides was reported [16]. Plasmid-borne extended-spectrum β-lactamases (ESBL), AmpC β-lactamases, and carbapenemases are the most concerning β-lactamases, since they confer resistance to broad-spectrum β-lactams. We described here eleven P. mirabilis clinical isolates producing NDM-1 carbapenemase. Of note, in addition to NDM-1 in all isolates, one isolate (P2-109-B1) carried the minor extended-spectrum lactamase blaVEB-6, a minor variant of blaVEB-4, which confers a high level of resistance to ceftazidime, cefotaxime, and aztreonam [34]. blaVEB-6 was first reported in a strain of P. mirabilis recovered from a urine sample of an inpatient in Australia [35]. Interestingly, co-localization of blaNDM-1 and blaVEB-6 in a same PGI (Proteus Genomic Island) has been reported in France [36]. As was observed for our 11 clinical isolates of P. mirabilis, the authors failed to transfer blaNDM-1 by the conjugation or electroporation in E. coli, despite demonstrating that the PGI was able to achieve self-excision and circularization, increasing the risk of co-transfer with another plasmid helper. This aligns with previous studies showing that integrative and conjugative elements (ICEs) and transposons play a crucial role in bacterial genome plasticity, allowing resistance determinants to persist in the chromosome while still being mobilized under specific conditions [37]. Furthermore, genomic islands with excision and integration capabilities have been implicated in the spread of antibiotic-resistant strains in healthcare settings, facilitating adaptation to selective pressures [38]. The inability to transfer blaNDM-1 in laboratory conditions does not rule out its dissemination in clinical settings, where helper elements such as conjugative plasmids or bacteriophage-mediated mechanisms may facilitate horizontal transfer, as reported in other carbapenemase-producing strains [39,40]. These findings highlight the complexity of resistance gene transmission and underscore the need for continuous surveillance of such elements in hospital environments.
While carbapenem-resistant Enterobacterales (CRE) have become a pressing threat to public health, current research in several countries has revealed that P. mirabilis, which produces carbapenemases, is still uncommon [41,42,43]. However, multidrug-resistant (MDR) P. mirabilis isolates harboring carbapenemase genes such as blaOXA-48-like, blaKPC, blaNDM, blaVIM, blaIMP, and the main CHDLs from A. baumannii, blaOXA-23 and blaOXA-58, are increasingly being reported [16,22,44,45]. They have frequently been described with co-resistance to fluoroquinolones, aminoglycosides, and co-trimoxazole [46,47,48].
Few cases of NDM-producing P. mirabilis have been described worldwide. In Tunisia, only one case reported by Kanzari et al. in 2018 [49] displaying the blaNDM-1 gene in an XDRPm clinical isolate carrying plasmid mediated resistance to carbapenems (blaNDM-1), cephalosporins (blaCMY-4), aminoglycosides (aph3-VIa and aph3-Ia), and fluoroquinolones (qnrA6) has been reported. Since that report, no other descriptions have been published in Tunisia.
The acquisition of blaNDM-1 is of special concern in relation to P. mirabilis, which is intrinsically resistant to tetracycline, tigecycline, and colistin. Indeed, this enzyme drastically diminishes the efficacy of almost all β-lactams (except aztreonam), including the last resort, carbapenems [23]. The extensive resistance observed in our P. mirabilis isolates severely limits treatment options. Notably, resistance to CAZ-AVI but susceptibility to ATM-AVI suggests that ceftazidime-avibactam combined with aztreonam may be a viable alternative against these NDM-1 producers [50].
Given the lack of effective β-lactams, cefiderocol remains a promising option due to its activity against NDM-producing pathogens [51]. These findings underscore the urgent need for antimicrobial stewardship, strict infection control, and continuous surveillance to contain the spread of multidrug-resistant P. mirabilis.
Beyond antimicrobial resistance, the pathogenicity of P. mirabilis is further enhanced by its virulence factors. The production of urease and protease contributes to stone formation and tissue necrosis, facilitating bacterial persistence in the host [8,52]. Additionally, all isolates carried hpmA and hpmB, which play a crucial role in hemolytic activity and urovirulence [53]. The presence of multiple flagellar genes (flgN, flhA, fliA, fliC, fliF, fliG, fliL, fliP) supports the bacterium’s motility and colonization abilities, despite the absence of flaD, which is not essential for swarming [26,54]. Key virulence factors such as ptA, zapA, mrpA, pmfA, mrpH, and atfA contribute to biofilm formation and increased antibiotic resistance, further complicating treatment [24,25,26,55]. Moreover, the utilization of phosphate transport and iron uptake systems enhances bacterial survival and pathogenic potential, emphasizing the multidimensional threat posed by these isolates [56,57].
This study reports the first outbreak of NDM-producing P. mirabilis in Africa, highlighting the emergence of this multidrug-resistant pathogen in Tunisia and its potential to spread within healthcare settings. Genomic analysis revealed evidence of clonal dissemination, suggesting nosocomial transmission and underscoring the urgent need for enhanced infection control measures. The resistance profile of these isolates, particularly their low MICs to ertapenem, raises concerns about the potential underestimation of carbapenem resistance in P. mirabilis. Further genomic and phenotypic studies are crucial, especially regarding OXA-23-producing strains with low carbapenem MICs, to better assess the clinical relevance and effectiveness of carbapenem therapy in such cases.
Beyond its clinical impact, the spread of NDM-producing P. mirabilis poses a serious public health threat, reinforcing the need for continuous surveillance, robust antimicrobial stewardship programs, and strict containment strategies to prevent the further dissemination of multidrug-resistant bacteria in hospital settings.
Author Contributions
Conceptualization, N.J., L.T., W.M. and R.A.B.; methodology, N.J., S.O., T.N. and R.A.B.; validation, D.G., L.D. and R.A.B.; investigation, N.J., S.O., L.T. and A.J.; resources, S.A., W.N., A.T., W.M. and T.N.; writing—original draft preparation, N.J. and R.A.B.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Assistance Publique-Hôpitaux de Paris, the Université Paris-Saclay, the Institut National de la Santé et de la Recherche Médicale (INSERM), and grants from the French National Research Agency for the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) (ANR-10-LABX-33). This work was also supported by the “PHC Utique” program of the French Ministry of Foreign Affairs, the French Ministry of Higher Education, Research and Innovation, and the Tunisian Ministry of Higher Education and Scientific Research under the CMCU project number 21G0807.
Institutional Review Board Statement
This study was reviewed and approved by the University Hospital of Sahloul—Sousse (Tunisia) Ethics Committee under the reference “HS09-2024” on 13 February 2024.
Informed Consent Statement
Our study does not require written informed consent, as it is a retrospective investigation at the University Hospital of Sahloul-Sousse, Tunisia, using a long-preserved strain collection. Ethical guidelines allow consent exemptions for studies without direct patient interaction, anonymized data, and pre-existing biological samples, ensuring no additional risk.
Data Availability Statement
The assembled genomes of strains from the current study have been deposited in GenBank under the accession numbers JAWLIO000000000, JAWMAP000000000, JAWKAW000000000, JAXFYQ000000000, JAWMTJ000000000, JAWMAQ000000000, JAWMAN000000000, JAWONU000000000, JAXFYP000000000, JAWLVG000000000, JAWMAO000000000.
Acknowledgments
We thank the many dedicated staff who have helped with data collection and technical issues: the staff of the Laboratory of Clinical Micobiology, the Department of Anesthesia and Intensive Care at the University Hospital Sahloul, Sousse, Tunisia and the members of the French National Reference Center for Antibiotic Resistance “Carbapenemase-producing Enterobacteriaceae. The authors would like to express their gratitude to the ESCMID Study Group for Antimicrobial Resistance Surveillance (ESGARS) for their valuable support and contributions to antimicrobial resistance research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Alanis, A.J. Resistance to Antibiotics: Are We in the Post-Antibiotic Era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [PubMed]
- Abban, M.K.; Ayerakwa, E.A.; Mosi, L.; Isawumi, A. The burden of hospital acquired infections and antimicrobial resistance. Heliyon 2023, 9, e20561. [Google Scholar] [PubMed]
- Lim, C.; Takahashi, E.; Hongsuwan, M.; Wuthiekanun, V.; Thamlikitkul, V.; Hinjoy, S.; Day, N.P.; Peacock, S.J.; Limmathurotsakul, D. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. eLife 2016, 5, e18082. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar]
- Jamil, R.T.; Foris, L.A.; Snowden, J. Proteus mirabilis Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK442017/ (accessed on 10 October 2023).
- Griffith, D.P.; Musher, D.M.; Itin, C. Urease. The primary cause of infection-induced urinary stones. Investig. Urol. 1976, 13, 346–350. [Google Scholar]
- Foxman, B.; Brown, P. Epidemiology of urinary tract infections: Transmission and risk factors, incidence, and costs. Infect. Dis. Clin. N. Am. 2003, 17, 227–241. [Google Scholar]
- Li, X.; Zhao, H.; Lockatell, C.V.; Drachenberg, C.B.; Johnson, D.E.; Mobley, H.L.T. Visualization of Proteus mirabilis within the Matrix of Urease-Induced Bladder Stones during Experimental Urinary Tract Infection. Infect. Immun. 2002, 70, 389–394. [Google Scholar] [PubMed]
- Norsworthy, A.N.; Pearson, M.M. From Catheter to Kidney Stone: The Uropathogenic Lifestyle of Proteus mirabilis. Trends Microbiol. 2017, 25, 304–315. [Google Scholar]
- Pearson, M.M.; Rasko, D.A.; Smith, S.N.; Mobley, H.L.T. Transcriptome of Swarming Proteus mirabilis. Infect. Immun. 2010, 78, 2834–2845. [Google Scholar]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and Urinary Tract Infections. Microbiol. Spectr. 2015, 3, 10.1128/microbiolspec.UTI-0017-2013. [Google Scholar] [CrossRef]
- Ullah, S.; Saadaat, R.; Hamidi, H.; Haidary, A.M. Proteus mirabilis: A rare cause of pneumonia, radiologically mimicking malignancy of the lung. Clin. Case Rep. 2023, 11, e7937. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.N.; Kim, N.J.; Kim, M.N.; Kim, Y.S.; Woo, J.H.; Ryu, J. Bacteraemia due to tribe Proteeae: A review of 132 cases during a decade (1991–2000). Scand. J. Infect. Dis. 2003, 35, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Endimiani, A.; Luzzaro, F.; Brigante, G.; Perilli, M.; Lombardi, G.; Amicosante, G.; Rossolini, G.M.; Toniolo, A. Proteus mirabilis bloodstream infections: Risk factors and treatment outcome related to the expression of extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 2005, 49, 2598–2605. [Google Scholar] [CrossRef]
- Stock, I. Natural Antibiotic Susceptibility of Proteus spp., with Special Reference to P. mirabilis and P. penneri Strains. J. Chemother. 2003, 15, 12–26. [Google Scholar] [PubMed]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. Front. Microbiol. 2020, 11, 256. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Hussein, A.I.A.; Shimamoto, T. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 2007, 59, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.S.; Mendonça, N.; Adeleke, O.E.; Da Silva, G.J. Molecular screening of antibiotic-resistant determinants among multidrug-resistant clinical isolates of Proteus mirabilis from SouthWest Nigeria. Afr. Health Sci. 2017, 17, 356. [Google Scholar] [CrossRef]
- Albornoz, E.; Lucero, C.; Romero, G.; Rapoport, M.; Guerriero, L.; Andres, P.; Group, W.A.; Galas, M.; Corso, A.; Petroni, A. Analysis of plasmid-mediated quinolone resistance genes in clinical isolates of the tribe Proteeae from Argentina: First report of qnrD in the Americas. J. Glob. Antimicrob. Resist. 2014, 2, 322–326. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis Infection. EcoSal Plus 2018, 8, 10.1128/ecosalplus.ESP-0009-2017. [Google Scholar] [CrossRef]
- Arpin, C.; Dubois, V.; Coulange, L.; André, C.; Fischer, I.; Noury, P.; Grobost, F.; Brochet, J.P.; Jullin, J.; Dutilh, B.; et al. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Community and Private Health Care Centers. Antimicrob. Agents Chemother. 2003, 47, 3506. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Girlich, D.; Jousset, A.B.; Gauthier, L.; Cuzon, G.; Bogaerts, P.; Haenni, M.; Madec, J.Y.; Couvé-Deacon, E.; Barraud, O.; et al. A single Proteus mirabilis lineage from human and animal sources: A hidden reservoir of OXA-23 or OXA-58 carbapenemases in Enterobacterales. Sci. Rep. 2020, 10, 9160. [Google Scholar]
- Usman Qamar, M.; SLopes, B.; Hassan, B.; Khurshid, M.; Shafique, M.; Atif Nisar, M.; Mohsin, M.; Nawaz, Z.; Muzammil, S.; Aslam, B.; et al. The present danger of New Delhi metallo-β-lactamase: A threat to public health. Future Microbiol. 2020, 15, 1759–1778. [Google Scholar]
- Beltrão, E.M.B.; Oliveira ÉM de Scavuzzi, A.M.L.; Firmo, E.F.; Lopes, A.C.d.S. Virulence factors of Proteus mirabilis clinical isolates carrying blaKPC-2 and blaNDM-1 and first report blaOXA-10 in Brazil. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2022, 28, 363–372. [Google Scholar]
- de Oliveira, W.D.; Barboza, M.G.; Faustino, G.; Inagaki, W.T.; Sanches, M.S.; Kobayashi, R.K.; Vespero, E.C.; Rocha, S.P. Virulence, resistance and clonality of Proteus mirabilis isolated from patients with community-acquired urinary tract infection (CA-UTI) in Brazil. Microb. Pathog. 2021, 152, 104642. [Google Scholar]
- Li, Y.; Yin, M.; Fang, C.; Fu, Y.; Dai, X.; Zeng, W.; Zhang, L. Genetic analysis of resistance and virulence characteristics of clinical multidrug-resistant Proteus mirabilis isolates. Front. Cell Infect. Microbiol. 2023, 13, 1229194. [Google Scholar]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.E.; Rolain, J.M.; Raoult, D. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar]
- EUCAST. Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 16 October 2023).
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid Detection of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar]
- Carba 5 Test—Carbapenemase Resistance Detection. Available online: https://www.ngbiotech.com/ng-test-carba-5/ (accessed on 24 October 2023).
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar]
- Kieser, T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 1984, 12, 19–36. [Google Scholar]
- Center for Genomic Epidemiology. Available online: http://genepi.food.dtu.dk/resfinder (accessed on 24 October 2023).
- Valentin, T.; Feierl, G.; Masoud-Landgraf, L.; Kohek, P.; Luxner, J.; Zarfel, G. Proteus mirabilis harboring carbapenemase NDM-5 and ESBL VEB-6 detected in Austria. Diagn. Microbiol. Infect. Dis. 2018, 91, 284–286. [Google Scholar]
- Zong, Z.; Partridge, S.R.; Iredell, J.R. RmtC 16S rRNA Methyltransferase in Australia. Antimicrob. Agents Chemother. 2008, 52, 794–795. [Google Scholar]
- Siebor, E.; De Curraize, C.; Neuwirth, C. Genomic context of resistance genes within a French clinical MDR Proteus mirabilis: Identification of the novel genomic resistance island GIPmi1. J. Antimicrob. Chemother. 2018, 73, 1808–1811. [Google Scholar] [PubMed]
- Botelho, J.; Schulenburg, H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2021, 29, 8–18. [Google Scholar] [PubMed]
- Wozniak, R.A.F.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [PubMed]
- Tansirichaiya, S.; Goodman, R.N.; Guo, X.; Bulgasim, I.; Samuelsen, Ø.; Al-Haroni, M.; Roberts, A.P. Intracellular Transposition and Capture of Mobile Genetic Elements following Intercellular Conjugation of Multidrug Resistance Conjugative Plasmids from Clinical Enterobacteriaceae Isolates. Microbiol. Spectr. 2022, 10, e0214021. [Google Scholar]
- Brown-Jaque, M.; Calero-Cáceres, W.; Muniesa, M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 2015, 79, 1–7. [Google Scholar]
- Horner, C.S.; Abberley, N.; Denton, M.; Wilcox, M.H. Surveillance of antibiotic susceptibility of Enterobacteriaceae isolated from urine samples collected from community patients in a large metropolitan area, 2010–2012. Epidemiol. Infect. 2014, 142, 399–403. [Google Scholar]
- Miró, E.; Agüero, J.; Larrosa, M.N.; Fernández, A.; Conejo, M.C.; Bou, G.; González-López, J.J.; Lara, N.; Martínez-Martínez, L.; Oliver, A.; et al. Prevalence and molecular epidemiology of acquired AmpC β-lactamases and carbapenemases in Enterobacteriaceae isolates from 35 hospitals in Spain. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2013, 32, 253–259. [Google Scholar]
- Sheng, W.H.; Badal, R.E.; Hsueh, P.R. Distribution of Extended-Spectrum β-Lactamases, AmpC β-Lactamases, and Carbapenemases among Enterobacteriaceae Isolates Causing Intra-Abdominal Infections in the Asia-Pacific Region: Results of the Study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob. Agents Chemother. 2013, 57, 2981–2988. [Google Scholar]
- Beltrão, E.M.B.; Oliveira, É.M.D.; Lopes, A.C.D.S. First report of bla GES-1 in Proteus mirabilis clinical isolates. Rev. Soc. Bras. Med. Trop. 2021, 54, e0864-2020. [Google Scholar]
- Hamprecht, A.; Sattler, J.; Noster, J.; Stelzer, Y.; Fuchs, F.; Dorth, V.; Gatermann, S.G.; Göttig, S. Proteus mirabilis—Analysis of a concealed source of carbapenemases and development of a diagnostic algorithm for detection. Clin. Microbiol. Infect. 2023, 29, 1198.e1–1198.e6. [Google Scholar] [PubMed]
- Korytny, A.; Riesenberg, K.; Saidel-Odes, L.; Schlaeffer, F.; Borer, A. Bloodstream infections caused by multi-drug resistant Proteus mirabilis: Epidemiology, risk factors and impact of multi-drug resistance. Infect. Dis. 2016, 48, 428–431. [Google Scholar]
- Lei, C.W.; Chen, Y.P.; Kang, Z.Z.; Kong, L.H.; Wang, H.N. Characterization of a Novel SXT/R391 Integrative and Conjugative Element Carrying cfr, bla CTX-M-65, fosA3, and aac(6′)-Ib-cr in Proteus mirabilis. Antimicrob. Agents Chemother. 2018, 62, e00849-18. [Google Scholar]
- Shaaban, M.; Elshaer, S.L.; Abd El-Rahman, O.A. Prevalence of extended-spectrum β-lactamases, AmpC, and carbapenemases in Proteus mirabilis clinical isolates. BMC Microbiol. 2022, 22, 247. [Google Scholar]
- Kanzari, L.; Ferjani, S.; Saidani, M.; Hamzaoui, Z.; Jendoubi, A.; Harbaoui, S.; Ferjani, A.; Rehaiem, A.; Boubaker, I.B.; Slim, A. First report of extensively-drug-resistant Proteus mirabilis isolate carrying plasmid-mediated blaNDM-1 in a Tunisian intensive care unit. Int. J. Antimicrob. Agents 2018, 52, 906–909. [Google Scholar]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar]
- McCreary, E.K.; Heil, E.L.; Tamma, P.D. New Perspectives on Antimicrobial Agents: Cefiderocol. Antimicrob. Agents Chemother. 2021, 65, e0217120. [Google Scholar]
- Alamuri, P.; Eaton, K.A.; Himpsl, S.D.; Smith, S.N.; Mobley, H.L.T. Vaccination with Proteus Toxic Agglutinin, a Hemolysin-Independent Cytotoxin In Vivo, Protects against Proteus mirabilis Urinary Tract Infection. Infect. Immun. 2009, 77, 632–641. [Google Scholar]
- Cestari, S.E.; Ludovico, M.S.; Martins, F.H.; da Rocha, S.P.D.; Elias, W.P.; Pelayo, J.S. Molecular detection of HpmA and HlyA hemolysin of uropathogenic Proteus mirabilis. Curr. Microbiol. 2013, 67, 703–707. [Google Scholar]
- Belas, R.; Suvanasuthi, R. The Ability of Proteus mirabilis To Sense Surfaces and Regulate Virulence Gene Expression Involves FliL, a Flagellar Basal Body Protein. J. Bacteriol. 2005, 187, 6789–6803. [Google Scholar]
- Belas, R.; Manos, J.; Suvanasuthi, R. Proteus mirabilis ZapA Metalloprotease Degrades a Broad Spectrum of Substrates, Including Antimicrobial Peptides. Infect. Immun. 2004, 72, 5159–5167. [Google Scholar] [PubMed]
- Mirzaei, A.; Habibi, M.; Bouzari, S.; Asadi Karam, M.R. Characterization of Antibiotic-Susceptibility Patterns, Virulence Factor Profiles and Clonal Relatedness in Proteus mirabilis Isolates from Patients with Urinary Tract Infection in Iran. Infect. Drug Resist. 2019, 12, 3967–3979. [Google Scholar] [PubMed]
- O’May, G.A.; Jacobsen, S.M.; Longwell, M.; Stoodley, P.; Mobley, H.L.T.; Shirtliff, M.E. The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology 2009, 155, 1523–1535. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).