The Global Outbreak of M. chimaera Infection Following Cardiac Surgery: Another Piece of the Puzzle
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Culture
2.3. Identification
2.4. Whole-Genome Sequencing
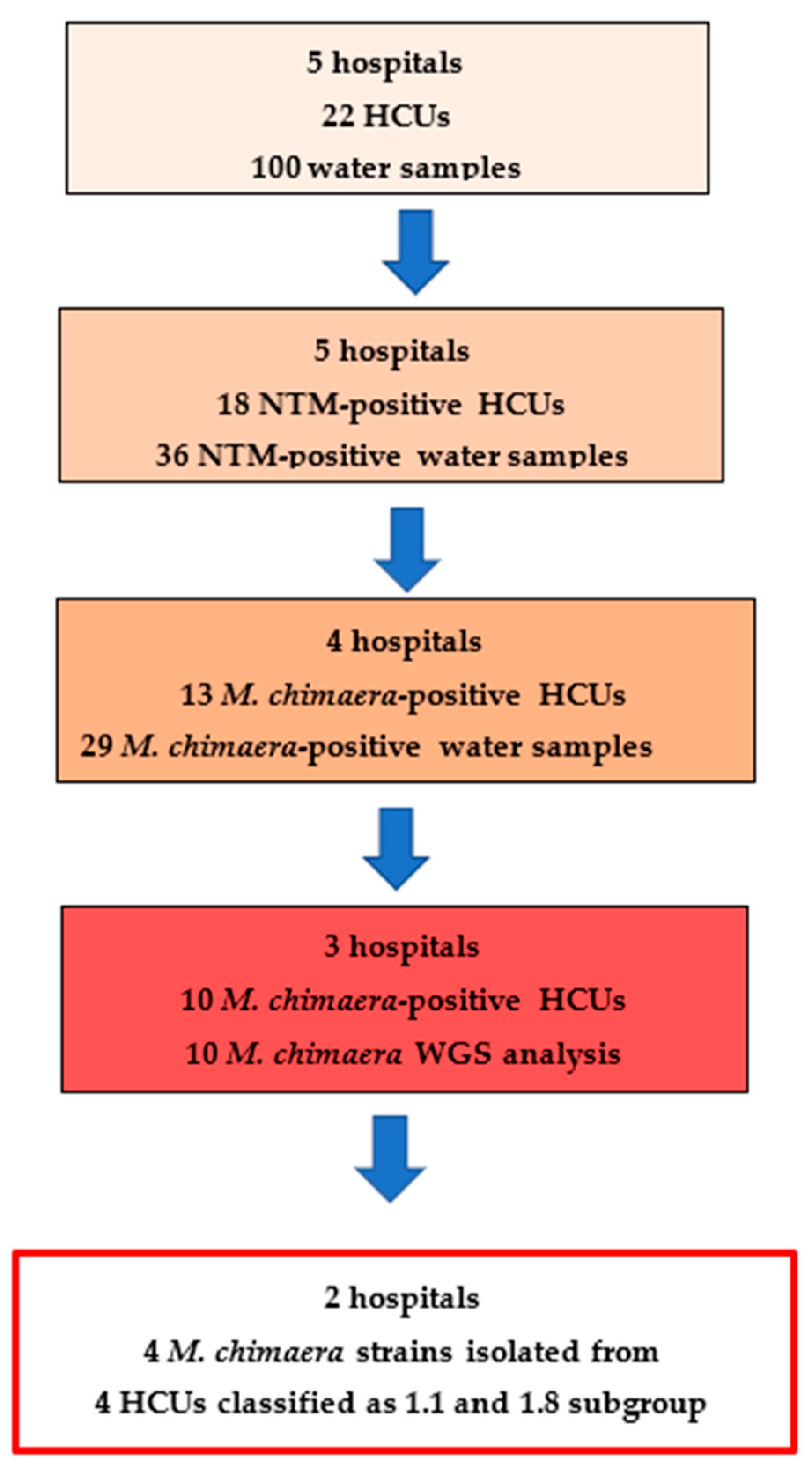
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Falkinham, J.O. Environmental Sources of Nontuberculous Mycobacteria. Clin. Chest Med. 2015, 36, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O.; Hilborn, E.D.; Arduino, M.J.; Pruden, A.; Edwards, M.A. Epidemiology and Ecology of Opportunistic Premise Plumbing Pathogens: Legionella Pneumophila, Mycobacterium Avium, and Pseudomonas Aeruginosa. Environ. Health Perspect. 2015, 123, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.M.; Gebert, M.J.; Delgado-Baquerizo, M.; Maestre, F.T.; Fierer, N. A Global Survey of Mycobacterial Diversity in Soil. Appl. Environ. Microbiol. 2019, 85, e01180-19. [Google Scholar] [CrossRef]
- Gebert, M.J.; Delgado-Baquerizo, M.; Oliverio, A.M.; Webster, T.M.; Nichols, L.M.; Honda, J.R.; Chan, E.D.; Adjemian, J.; Dunn, R.R.; Fierer, N. Ecological Analyses of Mycobacteria in Showerhead Biofilms and Their Relevance to Human Health. mBio 2018, 9, e01614-18. [Google Scholar] [CrossRef] [PubMed]
- CDC Clinical Overview of Nontuberculous Mycobacteria (NTM). Available online: https://www.cdc.gov/nontuberculous-mycobacteria/hcp/clinical-overview/index.html (accessed on 1 September 2025).
- U.S. Environmental Protection Agency. Legionella in the Indoor Environment. Available online: https://www.epa.gov/indoor-air-quality-iaq/legionella-indoor-environment (accessed on 1 September 2025).
- Li, T. A Systematic Review of Waterborne Infections from Nontuberculous Mycobacteria in Health Care Facility Water Systems. Int. J. Environ. Health 2017, 220, 611–620. [Google Scholar] [CrossRef]
- Honda, J.R.; Alper, S.; Bai, X.; Chan, E.D. Acquired and Genetic Host Susceptibility Factors and Microbial Pathogenic Factors That Predispose to Nontuberculous Mycobacterial Infections. Curr. Opin. Immunol. 2018, 54, 66–73. [Google Scholar] [CrossRef]
- Achermann, Y.; Rössle, M.; Hoffmann, M.; Deggim, V.; Kuster, S.; Zimmermann, D.R.; Bloemberg, G.; Hombach, M.; Hasse, B. Prosthetic Valve Endocarditis and Bloodstream Infection Due to Mycobacterium Chimaera. J. Clin. Microbiol. 2013, 51, 1769–1773. [Google Scholar] [CrossRef]
- Sax, H.; Bloemberg, G.; Hasse, B.; Sommerstein, R.; Kohler, P.; Achermann, Y.; Rössle, M.; Falk, V.; Kuster, S.P.; Böttger, E.C.; et al. Prolonged Outbreak of Mycobacterium chimaera Infection After Open-Chest Heart Surgery. Clin. Infect. Dis. 2015, 61, 67–75. [Google Scholar] [CrossRef]
- Haller, S.; Höller, C.; Jacobshagen, A.; Hamouda, O.; Abu Sin, M.; Monnet, D.L.; Plachouras, D.; Eckmanns, T. Contamination during Production of Heater-Cooler Units by Mycobacterium chimaera Potential Cause for Invasive Cardiovascular Infections: Results of an Outbreak Investigation in Germany, April 2015 to February 2016. Euro Surveill. 2016, 21, 1–6. [Google Scholar] [CrossRef]
- Sommerstein, R.; Rüegg, C.; Kohler, P.; Bloemberg, G.; Kuster, S.P.; Sax, H. Transmission of Mycobacterium chimaera from Heater-Cooler Units during Cardiac Surgery despite an Ultraclean Air Ventilation System. Emerg. Infect. Dis. 2016, 22, 1008–1013. [Google Scholar] [CrossRef]
- Chand, M.; Lamagni, T.; Kranzer, K.; Hedge, J.; Moore, G.; Parks, S.; Collins, S.; Del Ojo Elias, C.; Ahmed, N.; Brown, T.; et al. Insidious Risk of Severe Mycobacterium chimaera Infection in Cardiac Surgery Patients. Clin. Infect. Dis 2017, 64, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Perkins, K.M. Mycobacterium chimaera Contamination of Heater-Cooler Devices Used in Cardiac Surgery—United States. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1117–1118. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Finn, L.E.; Geist, R.L.; Prestel, C.; Moulton-Meissner, H.; Kim, M.; Stacey, B.; McAllister, G.A.; Gable, P.; Kamali, T.; et al. Mycobacterium chimaera Infections among Cardiothoracic Surgery Patients Associated with Heater-Cooler Devices-Kansas and California, 2019. Infect. Control Hosp. Epidemiol. 2021, 43, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Kohler, P.; Kuster, S.P.; Bloemberg, G.; Schulthess, B.; Frank, M.; Tanner, F.C.; Rössle, M.; Böni, C.; Falk, V.; Wilhelm, M.J.; et al. Healthcare-Associated Prosthetic Heart Valve, Aortic Vascular Graft, and Disseminated Mycobacterium chimaera Infections Subsequent to Open Heart Surgery. Eur. Heart J. 2015, 36, 2745–2753. [Google Scholar] [CrossRef]
- Bush, L.M.; Paturi, A.; Chaparro-Rojas, F.; Perez, M.T. Mycobacterial Prosthetic Valve Endocarditis. Curr. Infect. Dis. Rep. 2010, 12, 257–265. [Google Scholar] [CrossRef]
- Hasse, B.; Hannan, M.M.; Keller, P.M.; Maurer, F.P.; Sommerstein, R.; Mertz, D.; Wagner, D.; Fernández-Hidalgo, N.; Nomura, J.; Manfrin, V.; et al. International Society of Cardiovascular Infectious Diseases Guidelines for the Diagnosis, Treatment and Prevention of Disseminated Mycobacterium chimaera Infection Following Cardiac Surgery with Cardiopulmonary Bypass. J. Hosp. Infect. 2020, 104, 214–235. [Google Scholar] [CrossRef]
- van Ingen, J.; Kohl, T.A.; Kranzer, K.; Hasse, B.; Keller, P.M.; Katarzyna Szafrańska, A.; Hillemann, D.; Chand, M.; Schreiber, P.W.; Sommerstein, R.; et al. Global Outbreak of Severe Mycobacterium chimaera Disease after Cardiac Surgery: A Molecular Epidemiological Study. Lancet Infect. Dis. 2017, 17, 1033–1041. [Google Scholar] [CrossRef]
- ECDC. EU Protocol for Case Detection, Laboratory Diagnosis and Environmental Testing of Mycobacterium chimaera infections Potentially Associated with Heater-Cooler Units: Case Definition and Environmental Testing Methodology; ECDC: Stockholm, Sweden, 2015.
- Ministero della Salute. Indicazioni Operative Riguardanti Gli Aspetti Di Laboratorio in Merito Ai Casi Di Infezione Da Mycobacterium chimaera in Italia e Aggiornamento Delle Informazioni Disponibili; Ministero della Salute: Rome, Italy, 2019.
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Garlasco, J.; Zotti, C.M. Persistence of Legionella in Routinely Disinfected Heater-Cooler Units and Heater Units Assessed by Propidium Monoazide QPCR. Pathogens 2020, 9, 978. [Google Scholar] [CrossRef]
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Zotti, C.M. Real-Time PCR, the Best Approaches for Rapid Testing for Mycobacterium chimaera Detection in Heater Cooler Units and Extracorporeal Membrane Oxygenation. Perfusion 2021, 36, 626–629. [Google Scholar] [CrossRef]
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Garlasco, J.; Curtoni, A.; Iannaccone, M.; Zotti, C.M. A New Culture Method for the Detection of Non-Tuberculous Mycobacteria in Water Samples from Heater-Cooler Units and Extracorporeal Membrane Oxygenation Machines. Int. J. Environ. Res. Public Health 2022, 19, 10645. [Google Scholar] [CrossRef]
- Epperson, L.E.; Strong, M. A Scalable, Efficient, and Safe Method to Prepare High Quality DNA from Mycobacteria and Other Challenging Cells. J. Clin. Tuberc. Other Mycobact. Dis. 2020, 19, 100150. [Google Scholar] [CrossRef] [PubMed]
- Giacomuzzi, M.; Memoli, G.; Garlasco, J.; Zotti, C.M.; Ditommaso, S. Image Dataset of Environmental Non-Tuberculous Mycobacteria Isolated from Heater Cooler Units Water by the New NTM Elite Agar. Data Brief 2025, 59, 111358. [Google Scholar] [CrossRef] [PubMed]
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Cavallo, R.; Curtoni, A.; Avolio, M.; Silvestre, C.; Zotti, C.M. Reduction of Turnaround Time for Non-Tuberculous Mycobacteria Detection in Heater-Cooler Units by Propidium Monoazide-Real-Time Polymerase Chain Reaction. J. Hosp. Infect. 2019, 104, 365–373. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable Data Analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef]
- Cannas, A.; Campanale, A.; Minella, D.; Messina, F.; Butera, O.; Nisii, C.; Mazzarelli, A.; Fontana, C.; Lispi, L.; Maraglino, F.; et al. Epidemiological and Molecular Investigation of the Heater–Cooler Unit (HCU)-Related Outbreak of Invasive Mycobacterium chimaera Infection Occurred in Italy. Microorganisms 2023, 11, 2251. [Google Scholar] [CrossRef]
- Hasan, N.A.; Epperson, L.E.; Lawsin, A.; Rodger, R.R.; Perkins, K.M.; Halpin, A.L.; Perry, K.A.; Moulton-Meissner, H.; Diekema, D.J.; Crist, M.B.; et al. Genomic Analysis of Cardiac Surgery–Associated Mycobacterium chimaera Infections, United States. Emerg. Infect. Dis. 2019, 25, 559–563. [Google Scholar] [CrossRef]
- Svensson, E.; Jensen, E.T.; Rasmussen, E.M.; Folkvardsen, D.B.; Norman, A.; Lillebaek, T. Mycobacterium chimaera in Heater-Cooler Units in Denmark Related to Isolates from the United States and United Kingdom. Emerg. Infect. Dis. 2017, 23, 507–509. [Google Scholar] [CrossRef]
- Schreiber, P.W.; Sax, H. Mycobacterium chimaera Infections Associated with Heater-Cooler Units in Cardiac Surgery. Curr. Opin. Infect. Dis. 2017, 30, 388–394. [Google Scholar] [CrossRef]
- Williamson, D.; Howden, B.; Stinear, T. Mycobacterium chimaera Spread from Heating and Cooling Units in Heart Surgery. N. Engl. J. Med. 2017, 376, 600–602. [Google Scholar] [CrossRef]
- Ditommaso, S.; Garlasco, J.; Memoli, G.; Curtoni, A.; Bondi, A.; Ceccarelli, A.; Giacomuzzi, M. Emergence of Mycobacterium gordonae in Heater-Cooler Units: A Five-Year Prospective Surveillance of Devices Frequently Subjected to Chloramine-T Booster Disinfection. J. Hosp. Infect. 2024, 155, 9–16. [Google Scholar] [CrossRef]
| Sample ID | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MC-1180 | MC-830 | MC-988 | MC-1271B | MC-873 | MC-750 | MC-791 | MC-829 | MC-1345 | MC-872 | ||
|
Group/
Branch |
Associated
reference SNP position | ||||||||||
| Branch_1 | 4977262 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Branch_2 | 2339764 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Branch_2 | 5003561 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Group_1.1 | 113518 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Group_1.1 | 209278 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Group_1.11 | 1419163 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1.11 | 3132089 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1.6 | 4050336 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1.6 | 5033374 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1.8 *~ | 281696 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1.8 ~ | 1611282 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Group_1.8 ~ | 2366314 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Group_1 *~ | 2329494 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_1 *~ | 3949608 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_2.1 | 1828053 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_2.1 | 3406341 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_2 ~ | 3022332 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Group_2 ~ | 5709901 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sample Classification | Group 2 | Group 2 | Group 2 | Group 1.8 | Group 1.8 | Group 1.1 | Group 1.1 | Branch 2 | Un- grouped | Un grouped | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ditommaso, S.; Memoli, G.; Anselmi, F.; Molineris, I.; Zotti, C.M.; Giacomuzzi, M. The Global Outbreak of M. chimaera Infection Following Cardiac Surgery: Another Piece of the Puzzle. Pathogens 2025, 14, 964. https://doi.org/10.3390/pathogens14100964
Ditommaso S, Memoli G, Anselmi F, Molineris I, Zotti CM, Giacomuzzi M. The Global Outbreak of M. chimaera Infection Following Cardiac Surgery: Another Piece of the Puzzle. Pathogens. 2025; 14(10):964. https://doi.org/10.3390/pathogens14100964
Chicago/Turabian StyleDitommaso, Savina, Gabriele Memoli, Francesca Anselmi, Ivan Molineris, Carla Maria Zotti, and Monica Giacomuzzi. 2025. "The Global Outbreak of M. chimaera Infection Following Cardiac Surgery: Another Piece of the Puzzle" Pathogens 14, no. 10: 964. https://doi.org/10.3390/pathogens14100964
APA StyleDitommaso, S., Memoli, G., Anselmi, F., Molineris, I., Zotti, C. M., & Giacomuzzi, M. (2025). The Global Outbreak of M. chimaera Infection Following Cardiac Surgery: Another Piece of the Puzzle. Pathogens, 14(10), 964. https://doi.org/10.3390/pathogens14100964





