Multigene Typing of Croatian ‘Candidatus Phytoplasma Mali’ Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Insect Samples
2.2. MLST Primers and PCR Conditions
2.3. Sequence Analysis
3. Results
3.1. aceF, pnp, imp, and secY Genotyping
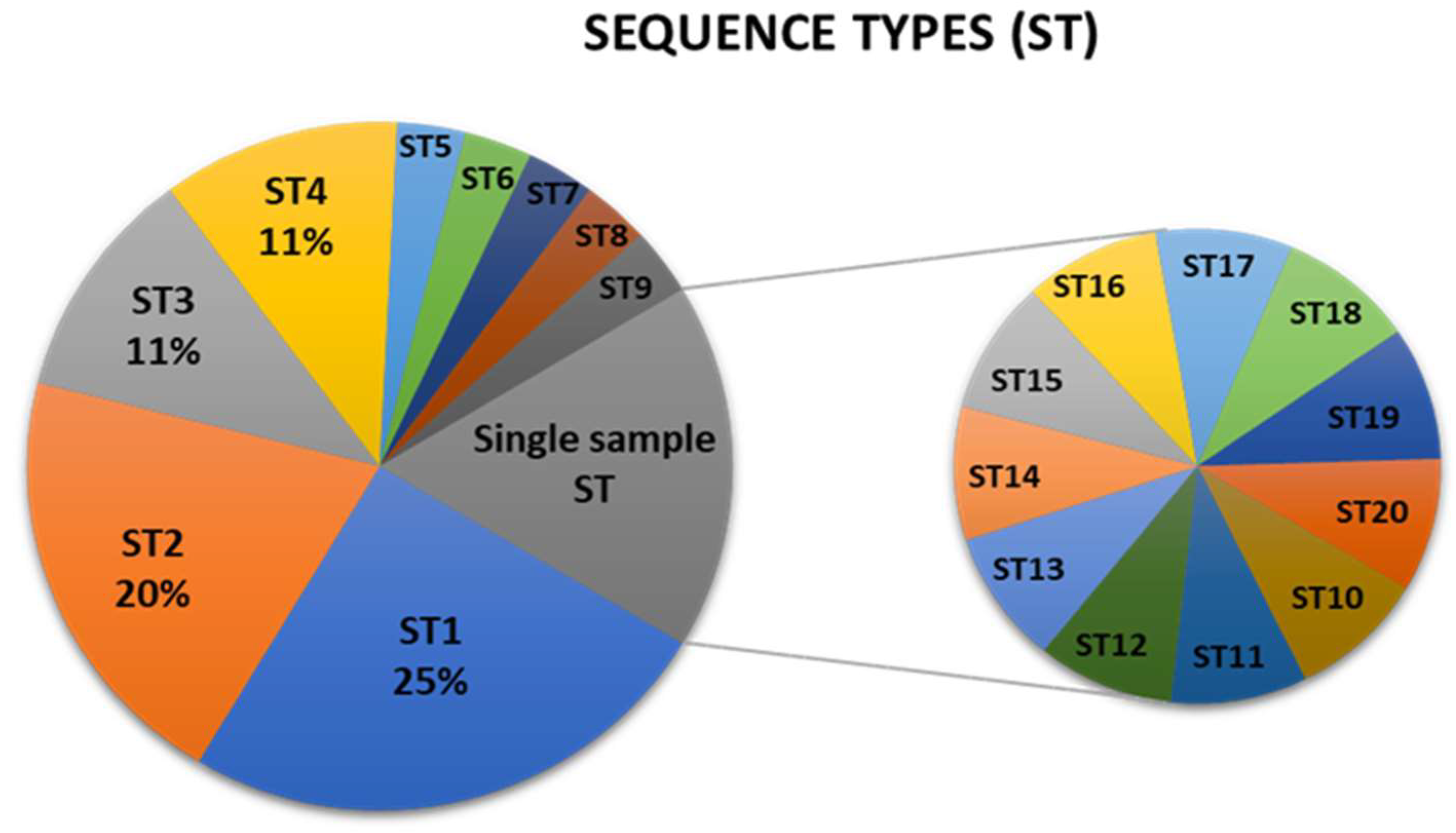
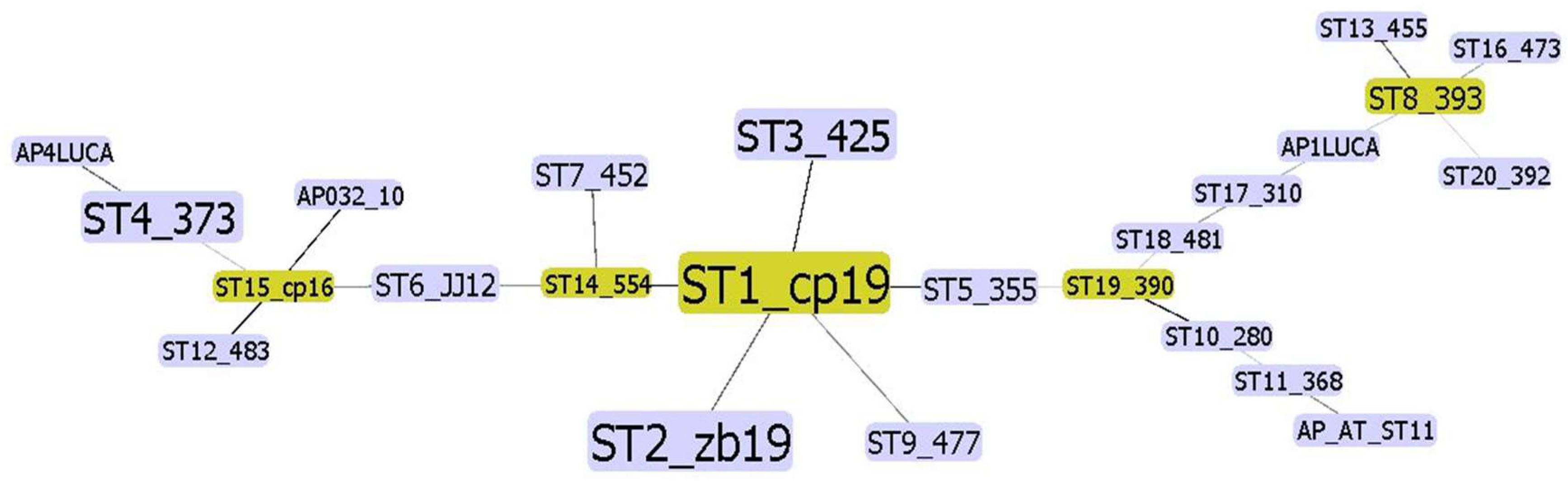
3.2. Sequence Types (STs) and Analysis of Concatenated Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Apple proliferation |
| MLST | Multi-locus sequence typing |
| ST | Sequence type |
References
- Marcone, C. Molecular Biology and Pathogenicity of Phytoplasmas. Ann. Appl. Biol 2014, 165, 199–221. [Google Scholar] [CrossRef]
- Lee, I.-M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic Mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef]
- IRPCM. ‘Candidatus Phytoplasma’, a Taxon for the Wall-Less, Non-Helical Prokaryotes That Colonize Plant Phloem and Insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhao, Y. Phytoplasma Taxonomy: Nomenclature, Classification, and Identification. Biology 2022, 11, 1119. [Google Scholar] [CrossRef] [PubMed]
- Kirdat, K.; Tiwarekar, B.; Sathe, S.; Yadav, A. From Sequences to Species: Charting the Phytoplasma Classification and Taxonomy in the Era of Taxogenomics. Front. Microbiol. 2023, 14, 1123783. [Google Scholar] [CrossRef]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.-Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.; Ugaki, M.; et al. Reductive Evolution Suggested from the Complete Genome Sequence of a Plant-Pathogenic Phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef]
- Seemüller, E.; Schneider, B. ‘Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’ and ‘Candidatus Phytoplasma prunorum’, the Causal Agents of Apple Proliferation, Pear Decline and European Stone Fruit Yellows, respectively. Int. J. Syst. Evol. Microbiol. 2004, 54, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The Linear Chromosome of the Plant-Pathogenic Mycoplasma “Candidatus Phytoplasma mali”. BMC Genomics 2008, 9, 306. [Google Scholar] [CrossRef]
- Urwin, R.; Maiden, M.C.J. Multi-Locus Sequence Typing: A Tool for Global Epidemiology. Trends Microbiol. 2003, 11, 479–487. [Google Scholar] [CrossRef]
- Danet, J.L.; Balakishiyeva, G.; Cimerman, A.; Sauvion, N.; Marie-Jeanne, V.; Labonne, G.; Laviňa, A.; Batlle, A.; Križanac, I.; Škorić, D.; et al. Multilocus Sequence Analysis Reveals the Genetic Diversity of European Fruit Tree Phytoplasmas and Supports the Existence of Inter-Species Recombination. Microbiology 2011, 157, 438–450. [Google Scholar] [CrossRef]
- Janik, K.; Barthel, D.; Oppedisano, T.; Anfora, G.; Schuler, H. Apple Proliferation—A Joint Review; Janik, K., Barthel, D., Oppedisano, T., Anfora, G., Eds.; Laimburg Research Centre: Stadio-Laimburg, Italy, 2020; ISBN 9788878430549. [Google Scholar]
- Šarić, A.; Cvjetković, B. Mycoplasma-like Organism Associated with Apple Proliferation and Pear Decline–like Disease of Pears. Agric. Conspec. Sci. 1985, 68, 61–65. [Google Scholar]
- Križanac, I.; Mikec, I.; Budinščak, Z.; Šeruga Musić, M.; Krajačić, M.; Škorić, D. Pomaceous Fruit Tree Phytoplasmas and Their Potential Vectors in Croatia. Acta Hortic. 2008, 781, 477–482. [Google Scholar] [CrossRef]
- Križanac, I.; Plavec, J.; Budinšćak, Ž.; Ivić, D.; Škorić, D.; Šeruga Musić, M. Apple Proliferation Disease in Croatian Orchards: A Molecular Characterization of “Candidatus Phytoplasma mali”. J. Plant Pathol. 2017, 99, 95–101. [Google Scholar]
- Frisinghelli, C.; Delaiti, L.; Grando, M.S.; Forti, D.; Vindimian, M.E. Cacopsylla Costalis (Flor 1861), as a Vector of Apple Proliferation in Trentino. J. Phytopath. 2000, 148, 425–431. [Google Scholar] [CrossRef]
- Tedeschi, R.; Bosco, D.; Alma, A. Population Dynamics of Cacopsylla melanoneura; (Homoptera: Psyllidae), a Vector of Apple Proliferation Phytoplasma in Northwestern Italy. J. Econ. Entomol. 2002, 95, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Budinšćak, Ž. Lisne Buhe–Vektori Fitoplazme Proliferacije Jabuke u Hrvatskoj. GBZ 2021, 21, 587–603. [Google Scholar]
- Baric, S.; Dalla-Via, J. A New Approach to Apple Proliferation Detection: A Highly Sensitive Real-Time PCR Assay. J. Microbiol. Methods 2004, 57, 135–145. [Google Scholar] [CrossRef]
- Deng, S.; Hiruki, C. Amplification of 16S RRNA Genes from Culturable and Nonculturable Mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Schneider, B.; Seemueller, E.; Smart, C.D.; Kirkpatrick, B.C. Phylogenetic Classification of Plant Pathogenic Mycoplasma-like Organisms or Phytoplasmas. In Molecular and Diagnostic Procedures in Mycoplasmology; Razin, S., Tully, J.G., Eds.; Elsevier: Amsterdam, The Netherlands, 1995; Volume 1, pp. 369–380. [Google Scholar]
- Lee, I.-M.; Gundersen, D.E.; Hammond, R.W.; Davis, R.E. Use of Mycoplasmalike Organism (MLO) Group-Specific Oligonucleotide Primers for Nested-PCR Assays to Detect Mixed-MLO Infections in a Single Host Plant. Phytopathology 1994, 84, 559–566. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Thompson, J. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic Inference and Data Visualization for Sequence Based Typing Methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Dermastia, M.; Dolanc, D.; Mlinar, P.; Mehle, N. Molecular Diversity of ‘Candidatus Phytoplasma mali’ and ‘Ca. P. prunorum’ in Orchards in Slovenia. Eur. J. Plant. Pathol. 2018, 152, 791–800. [Google Scholar] [CrossRef]
- Seemüller, E.; Kiss, E.; Sule, S.; Schneider, B. Multiple Infection of Apple Trees by Distinct Strains of ‘Candidatus Phytoplasma mali’ and Its Pathological Relevance. Phytopathology 2010, 100, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.J.; Jarausch, B.; Jarausch, W.; Jelkmann, W.; Vilcinskas, A.; Gross, J. Cacopsylla melanoneura has no Relevance as Vector of Apple Proliferation in Germany. Phytopathology 2009, 99, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Mittelberger, C.; Obkircher, L.; Oettl, S.; Oppedisano, T.; Pedrazzoli, F.; Panassiti, B.; Kerschbamer, C.; Anfora, G.; Janik, K. The Insect Vector Cacopsylla picta Vertically Transmits the Bacterium ‘Candidatus Phytoplasma mali’ to Its Progeny. Plant Pathol. 2017, 66, 1015–1021. [Google Scholar] [CrossRef]
- Jarausch, B.; Schwind, N.; Fuchs, A.; Jarausch, W. Characteristics of the Spread of Apple Proliferation by Its Vector Cacopsylla picta. Phytopathology 2011, 101, 1471–1480. [Google Scholar] [CrossRef]
- Kison, H.; Seemüller, E. Differences in Strain Virulence of the European Stone Fruit Yellows Phytoplasma and Susceptibility of Stone Fruit Trees on Various Rootstocks to This Pathogen. J. Phytopathol. 2001, 149, 533–541. [Google Scholar] [CrossRef]
- Konnerth, A.; Krczal, G.; Boonrod, K. Immunodominant Membrane Proteins of Phytoplasmas. Microbiology 2016, 162, 1267–1273. [Google Scholar] [CrossRef]
- Kakizawa, S.; Oshima, K.; Namba, S. Diversity and Functional Importance of Phytoplasma Membrane Proteins. Trends Microbiol. 2006, 14, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Barbara, D.J.; Morton, A.; Clark, M.F.; Davies, D.L. Immunodominant Membrane Proteins from Two Phytoplasmas in the Aster Yellows Clade (Chlorante Aster Yellows and Clover Phyllody) Are Highly Divergent in the Major Hydrophilic Region. Microbiology 2002, 148, 157–167. [Google Scholar] [CrossRef][Green Version]
- Bohunická, M.; Valentová, L.; Suchá, J.; Nečas, T.; Eichmeier, A.; Kiss, T.; Cmejla, R. Identification of 17 ‘Candidatus Phytoplasma pyri’ Genotypes Based on the Diversity of the Imp Gene Sequence. Plant Pathol. 2018, 67, 971–977. [Google Scholar] [CrossRef]
- Alessio, F.I.; Bongiorno, V.A.; Marcone, C.; Conci, L.R.; Fernandez, F.D. Genetic Diversity in Phytoplasmas from X-Disease Group Based in Analysis of IdpA and Imp Genes. Microorganisms 2025, 13, 1170. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect Vectors of Phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Rashidi, M.; Galetto, L.; Bosco, D.; Bulgarelli, A.; Vallino, M.; Veratti, F.; Marzachì, C. Role of the Major Antigenic Membrane Protein in Phytoplasma Transmission by Two Insect Vector Species. BMC Microbiol. 2015, 15, 193. [Google Scholar] [CrossRef]
- Lee, I.-M.; Bottner-Parker, K.D.; Zhao, Y.; Davis, R.E.; Harrison, N.A. Phylogenetic Analysis and Delineation of Phytoplasmas Based on SecY Gene Sequences. Int. J. Syst. Evol. Microbiol. 2010, 60, 2887–2897. [Google Scholar] [CrossRef]
- Seemüller, E.; Sule, S.; Kube, M.; Jelkmann, W.; Schneider, B. The AAA+ ATPases and HflB/FtsH Proteases of ‘Candidatus Phytoplasma mali’: Phylogenetic Diversity, Membrane Topology, and Relationship to Strain Virulence. MPMI 2013, 26, 367–376. [Google Scholar] [CrossRef]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.-D.; Kamoun, S.; Hogenhout, S.A. AY-WB Phytoplasma Secretes a Protein That Targets Plant Cell Nuclei. Mol. Plant-Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Lee, I.-M.; Bottner, K.D.; Zhao, Y.; Botti, S.; Bertaccini, A.; Harrison, N.A.; Carraro, L.; Marcone, C.; Khan, A.J.; et al. Ribosomal Protein Gene-Based Phylogeny for Finer Differentiation and Classification of Phytoplasmas. Int. J. Syst. Evol. Microbiol. 2007, 57, 2037–2051. [Google Scholar] [CrossRef] [PubMed]
- Fránová, J.; Koloniuk, I.; Lenz, O.; Sakalieva, D. Molecular diversity of “Candidatus Phytoplasma mali” strains associated with apple proliferation disease in Bulgarian germplasm collection. Folia Microbiol. 2019, 64, 373–382. [Google Scholar] [CrossRef] [PubMed]

| Gene | Method | Primer Name | Primer Sequence (5′–3′) | Amplicon Size (bp) | Coding Region (bp) |
|---|---|---|---|---|---|
| aceF * | PCR/ sequencing | acoB_F | CTGCTCCATCTAGAGTTAC | 1600 | 1260 |
| lpd_R0m | GCTAGCTTTTATAGCAGCT | ||||
| pnp | PCR | pnpF | GCTCAGTTGGTAGAGCAT | ||
| pnpR | AGACACAAACACTACATACAT | ||||
| Nested PCR/ sequencing | pnpF1 | GGTAGAGCATCTGACTGTT | 2400 | 2187 | |
| pnpR1 | CCCTCGATCGCCTTCTAT | ||||
| imp | PCR/ sequencing | impF | CGTAGAACCAAATGATAAAG | 1000 | 507 |
| impR | GACATAGACATCGTTACGA | ||||
| secY | PCR | secYF1 | CAGAGAATTCCTAAACGTG | ||
| secYR | GAATACCGTGAACAACTAC | ||||
| Nested PCR/ sequencing | secYF2 | GTTAATCTAGGTGCTTTAGA | 1800 | 1227–1248 | |
| secYR2 | GTGAACAACTACTTCATTAAC | ||||
| sequencing | secYF3 | AGTTGGTGGCAATATTGA | |||
| secYR3 | CTACTTCATTAACAGAAGTACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Križanac, I.; Šeruga Musić, M.; Plavec, J.; Škorić, D. Multigene Typing of Croatian ‘Candidatus Phytoplasma Mali’ Strains. Pathogens 2025, 14, 959. https://doi.org/10.3390/pathogens14100959
Križanac I, Šeruga Musić M, Plavec J, Škorić D. Multigene Typing of Croatian ‘Candidatus Phytoplasma Mali’ Strains. Pathogens. 2025; 14(10):959. https://doi.org/10.3390/pathogens14100959
Chicago/Turabian StyleKrižanac, Ivana, Martina Šeruga Musić, Jelena Plavec, and Dijana Škorić. 2025. "Multigene Typing of Croatian ‘Candidatus Phytoplasma Mali’ Strains" Pathogens 14, no. 10: 959. https://doi.org/10.3390/pathogens14100959
APA StyleKrižanac, I., Šeruga Musić, M., Plavec, J., & Škorić, D. (2025). Multigene Typing of Croatian ‘Candidatus Phytoplasma Mali’ Strains. Pathogens, 14(10), 959. https://doi.org/10.3390/pathogens14100959








