Multi-Marker Approach for the Identification of Different Heterodera Species (Nematoda: Heteroderidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Heterodera spp. Isolates
2.3. DNA Purification and Amplification
2.4. Sequencing
2.5. Heterodera spp. Phylogenetic Analysis
2.6. Phylogenetic Analysis
3. Results
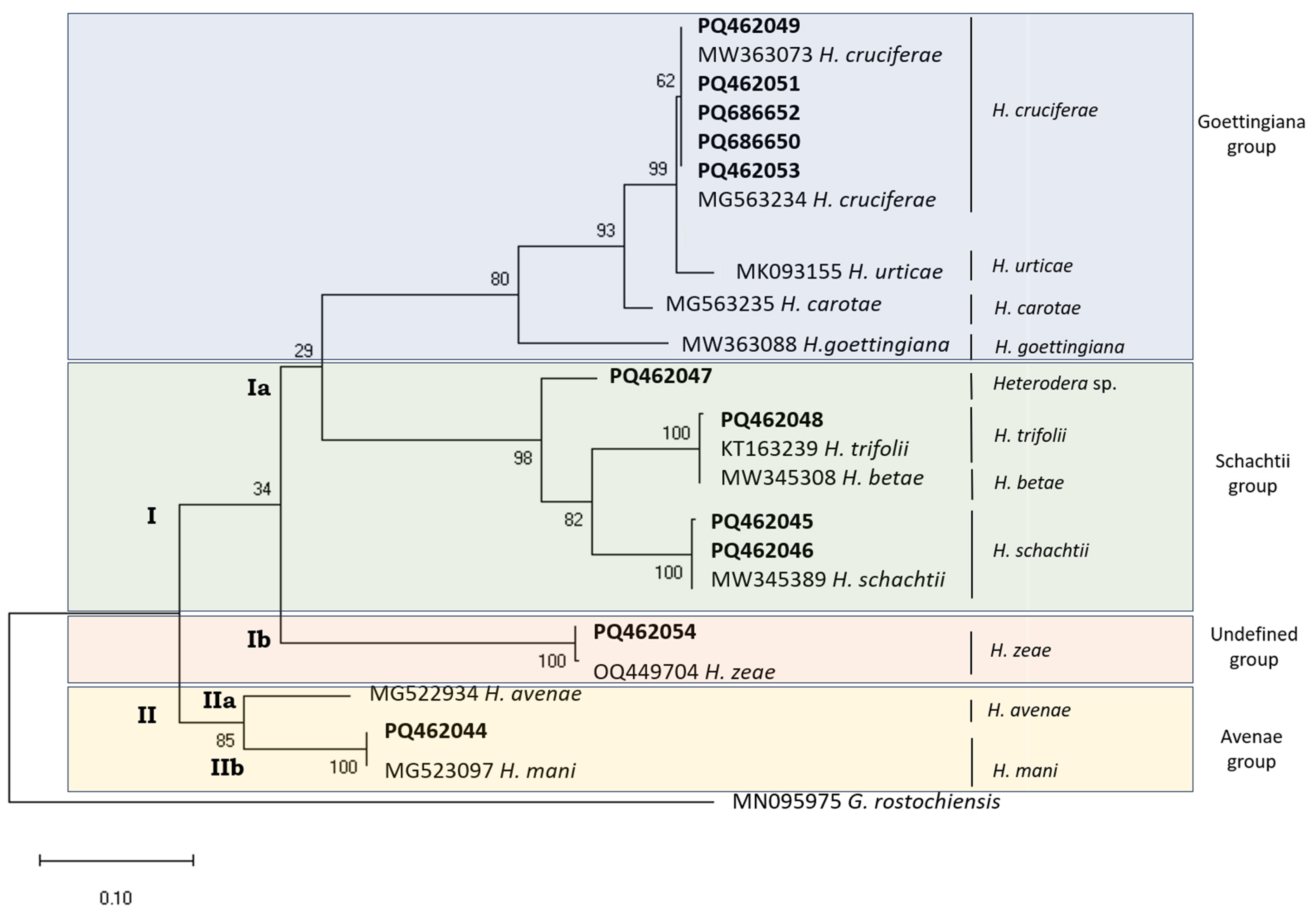
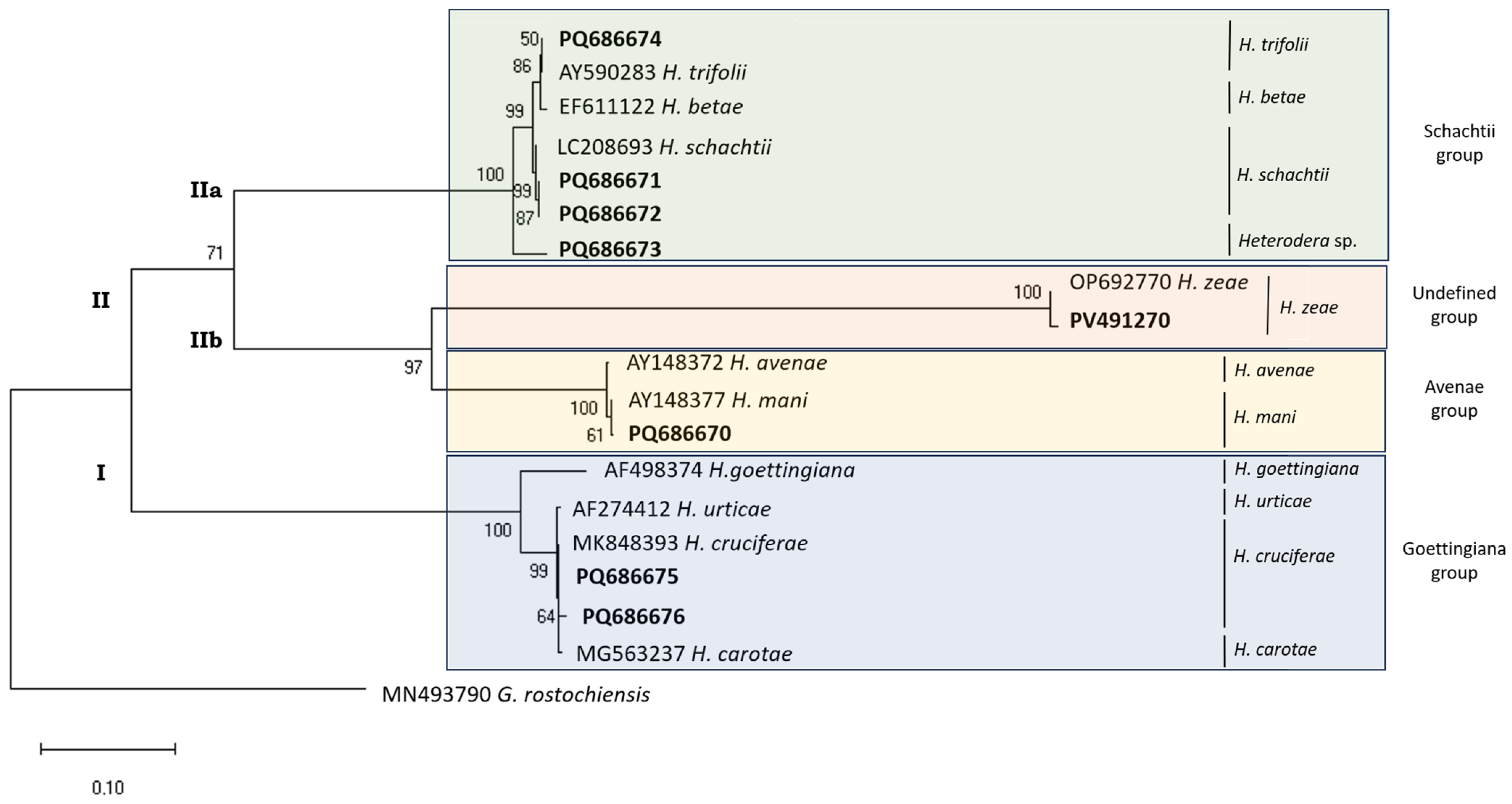
3.1. Partial mtCOI Gene Sequence
3.2. ITS Region
3.2.1. 18S rDNA
3.2.2. ITS-rDNA (18S rDNA–28S rDNA)
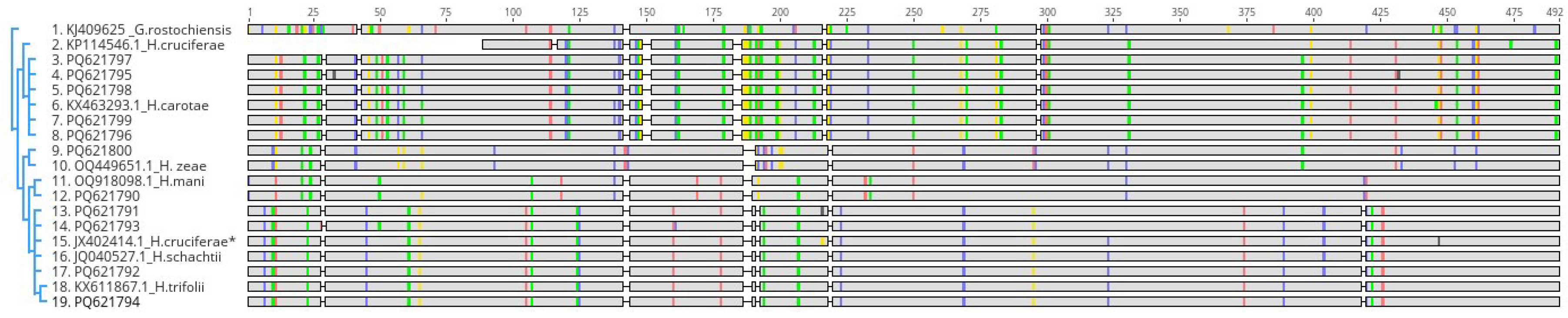
3.2.3. 28S rDNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huston, D.C.; Khudhir, M.; Hodda, M. Reliability and utility of standard gene sequence barcodes for the identification and differentiation of cyst nematodes of the genus Heterodera. J. Nematol. 2022, 54, 3041. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Mundo-Ocampo, M.; Baldwin, J.G. Systematics of Cyst Nematodes (Nematoda: Heteroderinae), Part B; Brill: Leiden, The Netherlands, 2010. [Google Scholar] [CrossRef]
- Azevedo, D. Impacte de Diferentes Culturas de Cobertura nos Nemátodes do Solo Num Campo na Golegã. Master’s Thesis, Instituto Superior de Agronomia, University of Lisbon, Lisbon, Portugal, 2024. Available online: https://repositorio.ulisboa.pt/bitstream/10400.5/99962/1/Diogo%20Azevedo_Disserta%c3%a7%c3%a3o.pdf (accessed on 6 May 2025).
- Turner, S.J.; Subbotin, S.A. Cyst nematodes. In Plant Nematology; CABI: Wallingford, UK, 2006; pp. 91–122. [Google Scholar]
- Waeyenberge, L.; Viaene, N.; Subbotin, S.; Moens, M. Molecular identification of Heterodera spp., an overview of fifteen years of research. In Cereal Cyst Nematodes: Status, Research and Outlook; Riley, I.T., Nicol, J.M., Dababat, A., Eds.; CIMMYT: Ankara, Turkey, 2009; pp. 109–114. [Google Scholar]
- European and Mediterranean Plant Protection Organization. PM 1/2 (32);EPPO A1 and A2 Lists of Pests Recommended for Regulations as Quarantine Pests; EPPO Standards; European and Mediterranean Plant Protection Organization: Paris, France, 2019; Available online: https://www.eppo.int/media/uploaded_images/ACTIVITIES/plant_quarantine/pm1-002-28-en.pdf (accessed on 7 September 2024).
- Szalanski, A.; Sui, D.D.; Harris, T.S.; Powers, T.O. Identification of cyst nematodes of agronomic and regulatory concern with PCR-RFLP of ITS1. J. Nematol. 1997, 29, 255–267. [Google Scholar]
- Subbotin, S.A.; Waeyenberge, L.; Moens, M. Identification of cyst forming nematodes of the genera Heterodera (Nematoda: Heteroderidae) based on the ribosomal DNA-RFLP. Nematology 2000, 2, 153–164. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Toumi, F.; Elekçioğlu, I.H.; Waeyenberge, L.; Maafi, Z.T. DNA barcoding, phylogeny and phylogeography of the cyst nematode species of the Avenae group from the genus Heterodera (Tylenchida: Heteroderidae). Nematology 2018, 20, 671–702. [Google Scholar] [CrossRef]
- Ferris, V.R.; Sabo, A.; Baldwin, J.G.; Mundo-Ocampo, M.; Inserra, R.N.; Sharma, S. Phylogenetic relationships among selected Heteroderoidea based on 18S and ITS ribosomal DNA. J. Nematol. 2004, 36, 202–206. [Google Scholar] [PubMed]
- Mundo-Ocampo, M.; Troccoli, A.; Subbotin, S.; Del Cid, J.; Baldwin, J.; Inserra, R. Synonymy of Afenestrata with Heterodera supported by phylogenetics with molecular and morphological characterisation of H. koreana comb. n. and H. orientalis comb. n. (Tylenchida: Heteroderidae). Nematology 2008, 10, 611–632. [Google Scholar] [CrossRef]
- Escobar-Avila, I.M.; López-Villegas, E.Ó.; Subbotin, S.A.; Tovar-Soto, A. First report of carrot cyst nematode Heterodera carotae in Mexico: Morphological, molecular characterization, and host range study. J. Nematol. 2018, 50, 229–242. [Google Scholar] [CrossRef]
- Powers, T.; Skantar, A.; Harris, T.; Higgins, R.; Mullin, P.; Hafez, S.; Handoo, Z.; Todd, T.; Powers, K. DNA barcoding evidence for the North American presence of alfalfa cyst nematode, Heterodera medicaginis. J. Nematol. 2019, 51, 1–17. [Google Scholar] [CrossRef]
- Ferris, V.R.; Ferris, J.M.; Faghihi, J. Variation in spacer ribosomal DNA in some cyst-forming species of plant parasitic nematodes. Fundam. Appl. Nematol. 1993, 16, 177–184. [Google Scholar]
- Subbotin, S.A.; Vierstraete, A.; De Ley, P.; Rowe, J.; Waeyenberge, L.; Moens, M.; Vanfleteren, J.R. Phylogenetic relationships within the cyst-forming nematodes (Nematoda, Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Mol. Phylogenetics Evol. 2001, 21, 1–16. [Google Scholar] [CrossRef]
- Vovlas, N.; Vovlas, A.; Leonetti, P.; Liébanas, G.; Castillo, P.; Subbotin, S.A.; Rius, J.E.P. Parasitism effects on white clover by root-knot and cyst nematodes and molecular separation of Heterodera daverti from H. trifolii. Eur. J. Plant Pathol. 2015, 143, 833–845. [Google Scholar] [CrossRef]
- Sekimoto, S.; Uehara, T.; Mizukubo, T. Characterisation of populations of Heterodera trifolii Goffart, 1932 (Nematoda: Heteroderidae) in Japan and their phylogenetic relationships with closely related species. Nematology 2017, 19, 543–558. [Google Scholar] [CrossRef]
- Reis, L.G.L. Occurrence of Cyst Nematodes in Portugal. In Cyst Nematodes; Lamberti, F., Taylor, C.E., Eds.; Nato ASI Series; Springer: Boston, MA, USA, 1986; Volume 121. [Google Scholar] [CrossRef]
- D’Oliveira, A.B. Nota sobre alguns nematodos de importância agricola. In Proceedings of the I Congresso Nacional das Ciencias Agrárias, Lisbon, Portugal, 24–31 October 1943; p. 117. 388p. [Google Scholar]
- Macara, A.M. Aspectos sobre a importância do nemátodos de interesse agricola em Portugal e no Ultramar português. Agros 1963, 46, 367–384. [Google Scholar]
- Macara, A.M. Contribuição para o estudo morfologico e biologico dos nematodos Heterodera goettingiana Liebscher, 1892 e rostochiensis Wollenweber, 1923 encontrados em Portugal. Broteria 1963, 32, 25. [Google Scholar]
- Macara, A.M. Contribuição Para o Estudo de Algumas Espécies do Género Heterodera Schmidt, 1871 Encontradas em Portugal. Ph.D. Thesis, Instituto Superior de Agronomia, Universidade Técnica de Lisboa, Lisbon, Portugal, 1962. [Google Scholar]
- Correia, F. Variabilidade Morfobiométrica de Populações de Nemátodes-dos-Quistos, Heterodera spp. Master’s Thesis, Universidade de Coimbra, Coimbra, Portugal, 1995. [Google Scholar]
- Madani, M.; Vovlas, N.; Castillo, P.; Subbotin, S.A.; Moens, M. Molecular Characterization of Cyst Nematode Species (Heterodera spp.) from the Mediterranean Basin using RFLPs and Sequences of ITS-rDNA. J. Phytopathol. 2004, 152, 229–234. [Google Scholar] [CrossRef]
- Vieira, P.; Sturhan, D.; Barbosa, P.; Padre, L.; Mota, M. List of the terrestrial nematodes (nematoda) from Azores. In A List of the Terrestrial and Marine Biota from the Azores; Borges, P.A.V., Costa, A., Cunha, R., Gabriel, R., Goncalves, V., Martins, A.F., Melo, I., Parente, M., Raposeiro, P., Rodrigues, P., et al., Eds.; Princípia: Cascais, Portugal, 2010. [Google Scholar]
- Gracianne, C.; Petit, E.J.; Arnaud, J.; Porte, C.; Renault, L.; Fouville, D.; Rouaux, C.; Fournet, S. Spatial distribution and basic ecology of Heterodera schachtii and H. betae wild populations developing on sea beet, Beta vulgaris ssp. maritima. Nematology 2014, 16, 797–805. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM 7/119 (1) Nematode extraction. EPPO Bull. 2013, 43, 471–495. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM 7/40 (5) Globodera rostochiensis and Globodera pallida. EPPO Bull. 2022, 52, 286–313. [Google Scholar] [CrossRef]
- Fenwick, D.W. Methods for the Recovery and Counting of Cysts of Heterodera schachtii from Soil. J. Helminthol. 1940, 18, 155–172. [Google Scholar] [CrossRef]
- Hu, M.; Hoglund, J.; Chilton, N.B.; Zhu, X.Q.; Gasser, R.B. Mutation scanning analysis of mitochondria cytochrome c oxidase subunit 1 reveals limited gene flow among bovine lungworm subpopulations in Sweden. Electrophoresis 2002, 23, 3357–3363. [Google Scholar] [CrossRef]
- Holterman, M.; Wurff, A.V.D.; Elsen, S.V.D.; Megen, H.V.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM 7/129 (2) DNA barcoding as an identification tool for a number of regulated pests. EPPO Bull. 2021, 51, 100–143. [Google Scholar] [CrossRef]
- De Ley, O.; Félix, M.A.; Frisse, L.; Nadler, S.; Sternberg, P.; Thomas, W. Molecular and morphological characterisation of two reproductively isolated species with mirrorimage anatomy (Nematoda: Cephalobidae). Nematology 1999, 1, 591–612. [Google Scholar] [CrossRef]
- Hall, T. Bioedit, version 7.0.9; 1997–2007; Ibis Biosciences: Carlsbad, CA, USA, 2007. [Google Scholar]
- Geneious Prime, version 2022.1; Biomatters Ltd.: Auckland, New Zealand, 2022; Available online: https://www.geneious.com (accessed on 9 June 2024).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J.; Clustal, W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]



| Heterodera Species | Locality (District/City) | Collection Code/Year | mtCOI Gene (Expected Frag. Size 447 bp) | 18S rDNA Gene (Frag. Size 1730 bp) | ITS rDNA Gene (Frag. Size 1040 bp) | 28S rDNA Gene (Frag. Size 780 bp) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GenBank Acc. No | bp | GenBank Acc. No. | bp | GenBank Acc. No | bp | GenBank Acc. No | bp | ||||
| H. cruciferae | Aveiro/Vagos | 1725 | 2021 | PQ462051 | 390 | PQ686668 | 1700 | PQ686676 | 975 | PQ621797 | 744 |
| H. cruciferae | Lisbon/Mafra | 1538-1 | 2020 | PQ462050 | 390 | PQ686667 | 1016 | PQ686675 | 984 | PQ621796 | 741 |
| H. cruciferae | Porto/Gondomar | 11391 | 2018 | PQ462049 | 390 | PQ686666 | 860 | PQ621795 | 740 | ||
| H. cruciferae | Santarém/Salvaterra | 15732 | 2018 | PQ462052 | 390 | PQ686669 | 1700 | PQ621798 | 718 | ||
| H. cruciferae | V. Real/Chaves | 2140 | 2020 | PQ462053 | 390 | PQ621799 | 740 | ||||
| H. mani | C. Branco/Covilhã | 13405 | 2018 | PQ462044 | 390 | PQ686661 | 1699 | PQ686670 | 978 | PQ621790 | 746 |
| H. schachtii | Faro/Loulé | 773-4 | 2020 | PQ462046 | 390 | PQ686663 | 1700 | PQ686672 | 964 | PQ621792 | 744 |
| H. schachtii | Leiria/Óbidos | 1856-1 | 2019 | PQ462045 | 390 | PQ686662 | 1700 | PQ686671 | 983 | PQ621791 | 745 |
| H. trifolii | V. Castelo/Melgaço | 1249-2 | 2019 | PQ462048 | 390 | PQ686665 | 1700 | PQ686674 | 962 | PQ621794 | 744 |
| H. zeae | Santarém/Golegã | 1978 | 2023 | PQ462054 | 390 | PV364147 | 1696 | PV491270 | 568 | PQ621800 | 737 |
| Heterodera sp. | Coimbra/Coimbra | 1086-1 | 2019 | PQ462047 | 390 | PQ686664 | 1702 | PQ686673 | 961 | PQ621793 | 744 |
| Heterodera Species | mtCOI Gene GenBank Accession Number | 18S rDNA Gene GenBank Accession Number | ITS rDNA Gene GenBank Accession Number | 28S rDNA Gene GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|---|
| H. avenae | MG522934 | 415 | KJ636290 | 1700 | AY148372 | 969 | LT159826 | 801 |
| H. betae | MW345308 | 434 | FJ040404 KJ636291 | 1700 1700 | EF611122 | 1027 | LC208670 | 737 |
| H. carotae | MG563235 | 424 | ** | MG563237 | 936 | KX463293 | 749 | |
| H. cruciferae | MG563234 MW363073 | 424 424 | AY566816 | 586 | MK848393 | 962 | KP114546 | 551 |
| H. cruciferae * | JX402414 | 747 | ||||||
| H. goettingiana | MW363088 | 424 | EU669915 | 1700 | AF498374 | 960 | DQ328697 | 653 |
| H. mani | MG523097 | 415 | EU669916 | 1700 | AY148377 | 968 | OQ918098 | 739 |
| H. schachtii | MW345389 | 434 | EU306355 | 1772 | LC208693 | 985 | JQ040527 | 780 |
| H. trifolii | KT163239 | 391 | FJ040402 | 1699 | AY590283 | 1011 | KX611867 | 754 |
| H. urticae | MK093155 | 868 | ** | AF274412 | 962 | DQ328696 | 653 | |
| H. zeae | OQ449704 | 391 | HQ724313 | 610 | OP692770 | 1033 | OQ449651 | 715 |
| G. rostochiensis | MN095975 | 443 | MZ613180 | 1740 | MN493790 | 973 | KJ409625 | 784 |
| Locality (District/City) | Marker | Isolate (Study) | Closest GenBank Match | Coverage | % Identity | Notes |
|---|---|---|---|---|---|---|
| Heterodera Cruciferae isolates | ||||||
| Aveiro/ Vagos | mtCOI | PQ462051 | H. cruciferae (MW363073) H. cruciferae (MG563234) H. urticae (MK093155I) H. carotae (MG563235) | 100% 100% 87% 100% | 100% 99.74% 99.71% 95.38% | Confirmed as H. cruciferae by mtCOI; only one H. urticae sequence available for comparison. ITS-rDNA (18L/ITS4mod) and 28S differentiates it from H. urticae. |
| 18S rDNA | PQ686668 | H. cruciferae (AY566816) H. goettingiana (EU669915) H. carotae H. carotae | 34% 100% No data No data | 99.83% 99.71% No data No data | Partial 18S rDNA similarity; only one H. cruciferae sequence available (586/1700 bp), while H. goettingiana sequence is complete, increasing misidentification risk. Identification confirmed by mtCOI and ITS-rDNA (18L/ITS4mod). This isolate adds a new database contribution. | |
| ITS | PQ686676 | H. cruciferae (MK848393) H. cruciferae (AF274411) H. carotae (MG563237) H. urticae (AF274412) H. goettingiana (AF498374) | 98% 98% 96% 98% 98% | 100% 99.90% 99.25% 98.85% 94.69% | Confirmed as H. cruciferae; identification supported by mtCOI, differs from H. carotae by mtCOI and from H. urticae by ITS-rDNA (18L/ITS4mod)—99% bootstrap value. | |
| 28S rDNA | PQ621797 | H. cruciferae (KP114546) H. urticae (DQ328696) H. carotae (KX463293) H. goettingiana (DQ328697) | 98% 91% 99% 91% | 98.90% 99.85% 99.44% 98.62% | Inconclusive for species identification; only one H. cruciferae sequence available (551/730 pb). However, Sequence clusters separately from H. carotae and H. urticae in NCBI phylogeny. This isolate adds a new database contribution. | |
| Lisbon/ Mafra | mtCOI | PQ462050 | H. cruciferae (MW363073) H. cruciferae (MG563234) H. urticae (MK093155I) H. carotae (MG563235) | 100% 100% 87% 100% | 100% 99.74% 99.71% 95.38% | Confirmed as H. cruciferae by mtCOI; only one H. urticae sequence available for comparison. ITS-rDNA (18L/ITS4mod) and 28S differentiates it from H. urticae. |
| 18S rDNA | PQ686667 | H. cruciferae (AY566816) H. goettingiana (EU669915) H. carotae H. carotae | 39% 100% No data No data | 99.75% 99.61% No data No data | Partial 18S rDNA similarity; only one H. cruciferae sequence available (586/1700 bp), while H. goettingiana sequence is complete, increasing misidentification risk. Identification confirmed by mtCOI and ITS-rDNA (18L/ITS4mod). This isolate adds a new database contribution. | |
| ITS | PQ686675 | H. cruciferae (MK848393) H. cruciferae (AF274411) H. carotae (MG563237) H. urticae (AF274412) H. goettingiana (AF498374) | 95% 98% 95% 98% 98% | 100% 99.90% 99.57% 99.17% 95.03% | Confirmed as H. cruciferae; identification supported by mtCOI, differs from H. carotae by mtCOI and from H. urticae by ITS-rDNA (18L/ITS4mod)—99% bootstrap value. | |
| 28S rDNA | PQ621796 | H. cruciferae (KP114546) H. urticae (DQ328696) H. carotae (KX463293) H. goettingiana (DQ328697) | 75% 98% 99% 88% | 99.10% 99.85% 99.59% 98.62% | Inconclusive for species identification; only one H. cruciferae sequence available (551/730 pb). However, sequence clusters separately from H. carotae and H. urticae in NCBI phylogeny. This isolate adds a new database contribution. | |
| Porto/ Gondomar | mtCOI | PQ462049 | H. cruciferae (MW363073) H. cruciferae (MG563234) H. urticae (MK093155I) H. carotae (MG563235) | 100% 100% 87% 100% | 100% 99.74% 99.71% 95.38% | Confirmed as H. cruciferae by mtCOI; only one H. urticae sequence available for comparison. ITS-rDNA (18L/ITS4mod) and 28S differentiates it from H. urticae. |
| 18S rDNA | PQ686666 | H. cruciferae (AY566816) H. goettingiana (EU669915) H. carotae H. carotae | 28% 100% No data No data | 98.35% 99.18% No data No data | Partial 18S rDNA similarity; only one H. cruciferae sequence available (586/1700 bp), while H. goettingiana sequence is complete, increasing misidentification risk. Identification confirmed by mtCOI and ITS-rDNA (18L/ITS4mod). This isolate adds a new database contribution. | |
| ITS | – | – | – | – | No amplification | |
| 28S rDNA | PQ621795 | H. cruciferae (KP114546) H. urticae (DQ328696) H. carotae (KX463293) H. goettingiana (DQ328697) | 75% 88% 99% 88% | 99.10% 99.69% 99.46% 98.16% | Inconclusive for species identification; only one H. cruciferae sequence available (551/730 pb). However, sequence clusters separately from H. carotae and H. urticae in NCBI phylogeny. This isolate adds a new database contribution. | |
| Santarém/ Salvaterra | mtCOI | PQ462052 | H. cruciferae (MW363073) H. cruciferae (MG563234) H. urticae (MK093155I) H. carotae (MG563235) | 100% 100% 87% 100% | 100% 99.74% 99.71% 95.38% | Confirmed as H. cruciferae by mtCOI; only one H. urticae sequence available for comparison. ITS-rDNA (18L/ITS4mod) and 28S differentiates it from H. urticae. |
| 18S rDNA | PQ686669 | H. cruciferae (AY566816) H. goettingiana (EU669915) H. carotae H. carotae | 34% 100% No data No data | 99.83% 99.71% No data No data | Partial 18S rDNA similarity; only one H. cruciferae sequence available (586/1700 bp), while H. goettingiana sequence is complete, increasing misidentification risk. Identification confirmed by mtCOI and ITS-rDNA (18L/ITS4mod). This isolate adds a new database contribution. | |
| ITS | – | – | – | – | No amplification | |
| 28S rDNA | PQ621798 | H. cruciferae (KP114546) H. urticae (DQ328696) H. carotae (KX463293) H. goettingiana (DQ328697) | 75% 91% 99% 91% | 98.9% 99.85% 99.44% 98.62% | Inconclusive for species identification; only one H. cruciferae sequence available (551/730 pb). However, sequence clusters separately from H. carotae and H. urticae in NCBI phylogeny. This isolate adds a new database contribution. | |
| V. Real/ Chaves | mtCOI | PQ462053 | H. cruciferae (MW363073) H. cruciferae (MG563234) H. urticae (MK093155I) H. carotae (MG563235) | 100% 100% 87% 100% | 99.74% 100% 100% 95.90% | Confirmed as H. cruciferae by mtCOI; only one H. urticae sequence available for comparison. ITS-rDNA (18L/ITS4mod) and 28S differentiates it from H. urticae. |
| 18S rDNA | – | – | – | – | No amplification | |
| ITS | – | – | – | – | No amplification | |
| 28S rDNA | PQ621799 | H. cruciferae (KP114546) H. urticae (DQ328696) H. carotae (KX463293) H. goettingiana (DQ328697) | 75% 88% 99% 88% | 99.28% 99.58% 99.86% 98.32% | Inconclusive for species identification; only one H. cruciferae sequence available (551/730 pb). However, sequence clusters separately from H. carotae and H. urticae in NCBI phylogeny. This isolate adds a new database contribution. | |
| Heterodera mani isolates | ||||||
| C. Branco/ Covilhã | mtCOI | PQ462044 | H. mani (MG523097) H. avenae (MG522934) | 100% 98% | 100% 90.86% | Confirmed as H. mani by mtCOI. |
| 18S rDNA | PQ686661 | H. mani (EU669916) H. aveane (KJ636290) | 100% 100% | 99.94% 99.88% | Only one H. mani sequence is currently available for comparison. Species identification is based on mtCOI and ITS-rDNA. This isolate provides a new contribution to the database. | |
| ITS | PQ686670 | H. mani (AY148377) H. avenae (AY148372) | 95% 99% | 99.69% 99.28% | Species identification is based on mtCOI. The H. mani sequences differ from H. avenae, with strong support (100% bootstrap value). | |
| 28S rDNA | PQ621790 | H. mani (OQ918098) H. avenae (LT159826) | 99% 100% | 99.86% 99.87% | Species identification is based on mtCOI and ITS-rDNA. | |
| Heterodera schachtii isolates | ||||||
| Faro/ Loulé | mtCOI | PQ462046 | H. schachtii (MW345380) H. schachtii (MW345389) | 100% 100% | 100% 100% | Confirmed as H. schachtiii by mtCOI. |
| 18S rDNA | PQ686663 | H. schachtii (EU306355) H. schachtii (KJ636284) H. trifolii (FJ040402) H. trifolii (KJ636287) H. betae (KJ636291) H. betae (FJ040404) | 100% 100% 100% 100% 100% 100% | 99.94% 99.88% 99.82% 99.76% 99.94% 99.94% | Inconclusive for species identification; species identification is based on mtCOI and ITS-rDNA (18L/ITS4mod), which clearly separate H. schachtii from both H. betae and H. trifolii. | |
| ITS | PQ686672 | H. schachtii (LC208693) H. trifolii (AY590283) H. betae (EF611122) | 100% 100% 100% | 99.9% 98.96% 98.45% | Confirmed as H. schachtii; ITS-rDNA is highly variable, enabling species-level identification with strong support (99% bootstrap). | |
| 28S rDNA | PQ621792 | H. schachtii (JQ040527) H. schachtii (MK895555) H. trifolii (KX611867) | 100% 100% 100% | 100% 100% 99.87% | 28S-rDNA has limited resolution, as H. schachtii differs from H. betae by 1 bp and from H. trifolii by 2 bp. Species identification relies on mtCOI and ITS-rDNA (18L/ITS4mod), which clearly separate H. schachtii from both H. betae and H. trifolii. | |
| Leiria/ Óbidos | mtCOI | PQ462045 | H. schachtii (MW345380) H. schachtii (MW345389) | 100% 100% | 99.74% 99.74% | Confirmed as H. schachtii by mtCOI. |
| 18S rDNA | PQ686662 | H. schachtii (EU306355) H. schachtii (KJ636284) H. trifolii (FJ040402) H. trifolii (KJ636287) H. betae (KJ636291) H. betae (FJ040404) | 100% 100% 100% 100% 100% 100% | 100% 99.94% 99.88% 99.82% 99.94% 100% | Inconclusive for species identification; species identification is based on mtCOI and ITS-rDNA (18L/ITS4mod), which clearly separate H. schachtii from both H. betae and H. trifolii. | |
| ITS | PQ686671 | H. schachtii (LC208693) H. trifolii (AY590283) H. betae (EF611122) | 99% 99% 99% | 99.48% 98.76% 98.25% | Confirmed as H. schachtii; ITS-rDNA is highly variable, enabling species-level identification with strong support (99% bootstrap). The sequences in this gene region are highly variable, allowing for the identification of isolates as H. schachtii, highly supported by a 99% bootstrap value | |
| 28S rDNA | PQ621791 | H. schachtii (JQ040527) H. schachtii (MK895555) H. trifolii (KX611867) | 100% 100% 100% | 99.60% 99.60% 99.46% | 28S-rDNA has limited resolution, as H. schachtii differs from H. betae by 1 bp and from H. trifolii by 2 bp. Species identification relies on mtCOI and ITS-rDNA (18L/ITS4mod), which clearly separate H. schachtii from both H. betae and H. trifolii. | |
| Heterodera trifolii isolates | ||||||
| V. Castelo/ Melgaço | mtCOI | PQ462048 | H. trifolii (KT163239) H. trifolii (MK621902) H. betae (MW345308) H. betae (LC208706) | 100% 100% 100% 100% | 99.74% 99.74% 99.74% 99.74% | mtCOI ambiguous (1 bp difference, low resolution); species identification confirmed by ITS-rDNA (18L/ITS4mod). |
| 18S rDNA | PQ686665 | H. schachtii (EU306355) H. betae (FJ040404) H. betae (KJ636291) H. trifolii (FJ040402) H. trifolii (KJ636287) | 100% 100% 100% 100% 100% | 100% 100% 99.94% 99.88% 99.82% | 18S rDNA ambiguous (1–2 bp differences; ITS regions shared among species); identification confirmed by ITS-rDNA (18L/ITS4mod) | |
| ITS | PQ686674 | H. trifolii (AY590283) H. betae (EF611122) H. schachtii (LC208693) | 100% 100% 100% | 100% 99.27% 99.06% | ITS-rDNA confirms H. trifolii; distinguished from H. betae by 7 nucleotide differences. | |
| 28S rDNA | PQ621794 | H. trifolii (KX611867) H. betae (LC208670) H. schachtii (MK895555) | 100% 99% 100% | 100% 99.86% 99.87% | 28S-rDNA shows low resolution (H. schachtii vs. H. betae differ by 1 bp; vs. H. trifolii by 2 bp); identification confirmed by ITS-rDNA (18L/ITS4mod) | |
| Heterodera zeae isolates | ||||||
| Santarém/ Golegã | mtCOI | PQ462054 | H. zeae (OQ449704) | 93% | 100% | Confirmed species |
| 18S rDNA | PV364147 | H. zeae (HQ724313) H. mani (EU669916) H. avenae (KJ636290) | 36% 100% 100% | 99.84% 98.53% 98.47% | Species identification is based on mtCOI and 28S. | |
| ITS | PV491270 | H. zeae (OP692769) H. zeae (OP692770) | 100% 100% | 99.12% 98.95% | Species identification is based on mtCOI and 28S. | |
| 28S rDNA | PQ621800 | H. zeae (OQ449651) H. zeae (GU145612) | 96% 100% | 100% 99.86% | Confirmed species | |
| Heterodera sp. isolates | ||||||
| Coimbra/ Coimbra | mtCOI | PQ462047 | H. sonchophila (MW345341) H. glycines (LC208712) H. medicaginis (MW345339) | 99% 99% 99% | 93.25% 92.99% 92.99% | Forms a distinct sub-clade; identification ambiguous, possibly representing an undescribed species. |
| 18S rDNA | PQ686664 | H. betae (FJ040404) H. schachtii (EU306355) H. schachtii (KJ636284) H. betae (KJ934138) H. hordecalis (FJ040405) | 100% 100% 100% 100% 100% | 99.72% 99.72% 99.65% 99.65% 99.65% | Forms a distinct sub-clade; identification ambiguous, possibly representing an undescribed species. | |
| ITS | PQ686673 | H. schachtii (EF611116) H. trifolii (AY590283) H. betae (LC208690) | 100% 100% 100% | 96.48% 96.37% 95.85% | Unique sub-clade; ambiguous; potentially undescribed species | |
| 28S rDNA | PQ621793 | H. schachtii (MK895554) H. schachtii (JQ040527) H. glycines (KP324916) H. trifolii (KX611876) | 100% 100% 100% 100% | 99.46% 99.33% 99.19% 99.19% | Unique sub-clade; ambiguous; potentially undescribed species | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camacho, M.J.; Inácio, M.L.; Andrade, E.d. Multi-Marker Approach for the Identification of Different Heterodera Species (Nematoda: Heteroderidae). Pathogens 2025, 14, 1052. https://doi.org/10.3390/pathogens14101052
Camacho MJ, Inácio ML, Andrade Ed. Multi-Marker Approach for the Identification of Different Heterodera Species (Nematoda: Heteroderidae). Pathogens. 2025; 14(10):1052. https://doi.org/10.3390/pathogens14101052
Chicago/Turabian StyleCamacho, Maria João, Maria L. Inácio, and Eugénia de Andrade. 2025. "Multi-Marker Approach for the Identification of Different Heterodera Species (Nematoda: Heteroderidae)" Pathogens 14, no. 10: 1052. https://doi.org/10.3390/pathogens14101052
APA StyleCamacho, M. J., Inácio, M. L., & Andrade, E. d. (2025). Multi-Marker Approach for the Identification of Different Heterodera Species (Nematoda: Heteroderidae). Pathogens, 14(10), 1052. https://doi.org/10.3390/pathogens14101052






