Potential Strategies Applied by Metschnikowia bicuspidata to Survive the Immunity of Its Crustacean Hosts
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strain and Cell Collection
2.2. Collection of Hemocytes
2.3. Sampling of Yeast Cells for Total RNA Isolation
2.4. RNA-Seq Analysis
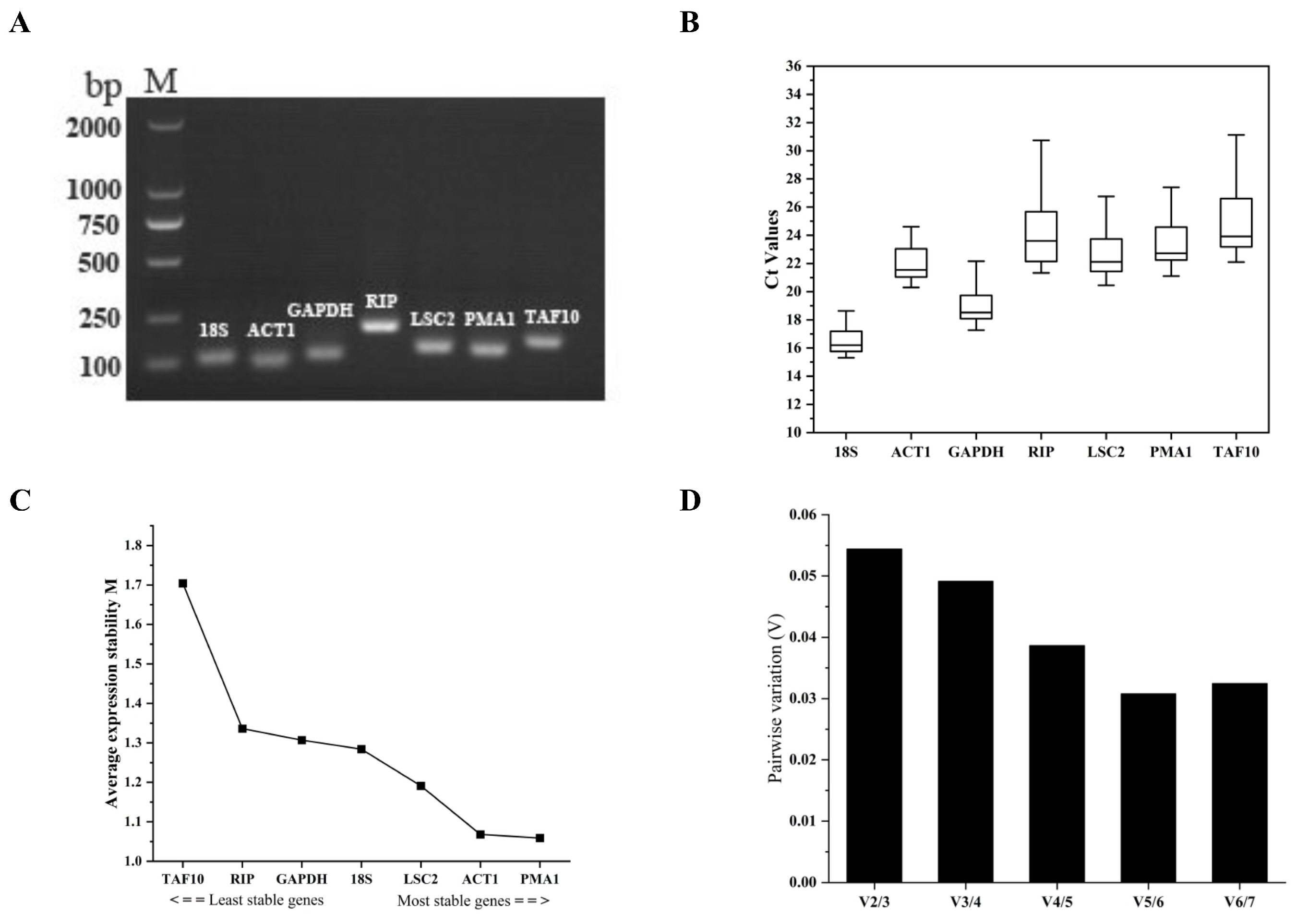
2.5. Reference Gene Evaluation
2.6. qRT-PCR Verification of the RNA-Seq Results
3. Results
3.1. Evaluation of the RNA-Seq Data
3.2. Initial Classification of the DEGs
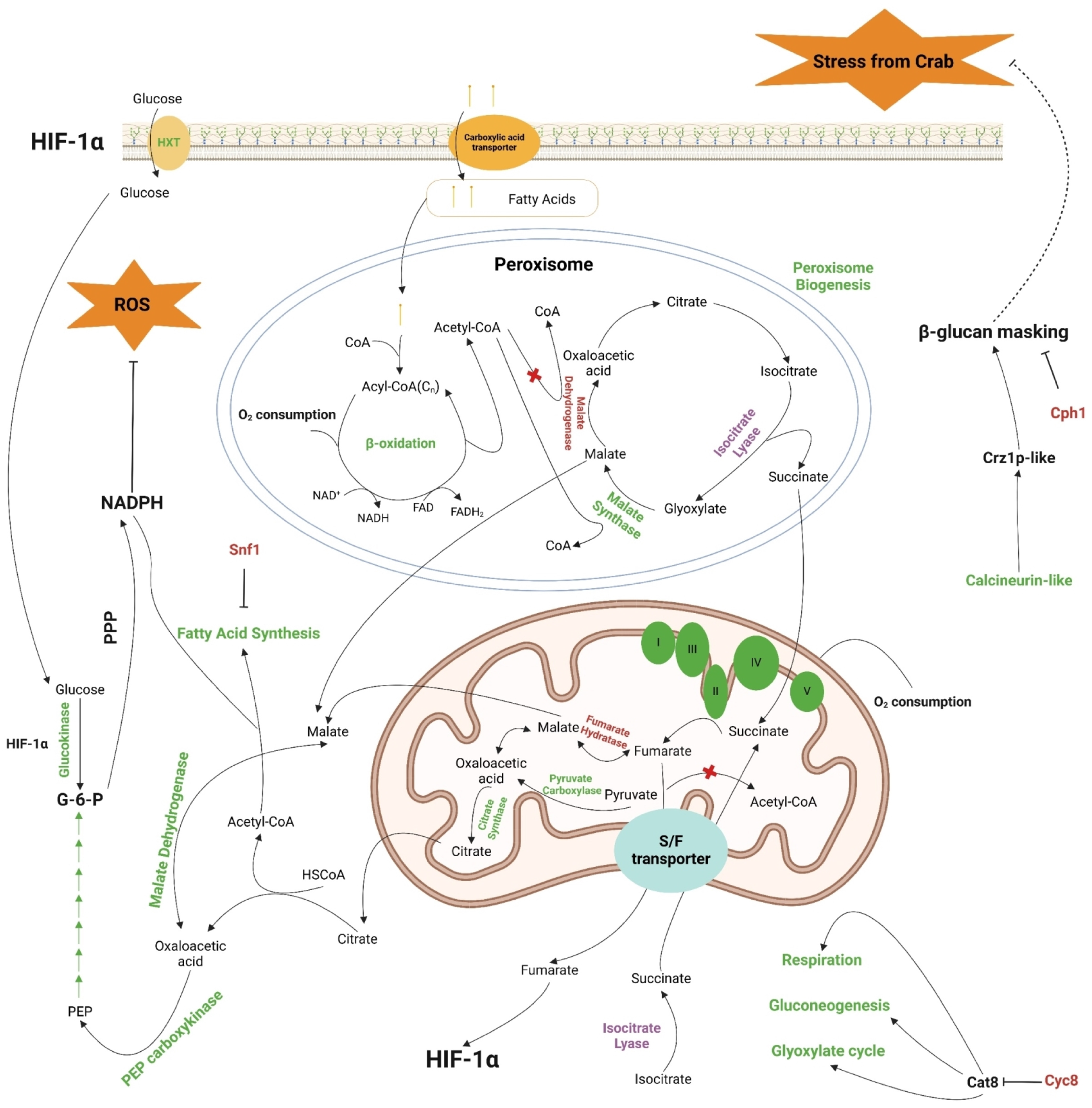
3.3. A Glance of the Most-Affected DEGs
3.4. Specificity and Amplification Efficiency of the Primer Pairs for Their Respective Candidate Reference Genes
3.5. Analyzing the Qualifications of Candidate Reference Genes
3.6. qRT-PCR Validation of the Transcriptomic Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metschnikoff, E. A disease of Daphnia caused by a yeast. A contribution to the theory of phagocytes as agents for attack on disease-causing organisms. Archiv. Pathol. Anat. Physiol. Klin. Med. 1884, 96, 177–195. [Google Scholar] [CrossRef]
- Lachance, M.-A. Metschnikowia Kamienski (1899). In The Yeasts; Elsevier: London, UK, 2011; pp. 575–620. [Google Scholar]
- Dallas, T.; Holtackers, M.; Drake, J.M. Costs of resistance and infection by a generalist pathogen. Ecol. Evol. 2016, 6, 1737–1744. [Google Scholar] [CrossRef]
- Cuco, A.P.; Castro, B.B.; Gonçalves, F.; Wolinska, J.; Abrantes, N. Temperature modulates the interaction between fungicide pollution and disease: Evidence from a Daphnia-microparasitic yeast model. Parasitology 2018, 145, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Lachance, M.-A.; Miranda, M.; Miller, M.W.; Phaff, H.J. Dehiscence and active spore release in pathogenic strains of the yeast Metschnikowia bicuspidata var. australis: Possible predatory implication. Can. J. Microbiol. 1976, 22, 1756–1761. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Chen, Y.-C.; Kwang, J.; Manopo, I.; Wang, P.-C.; Chaung, H.-C.; Liaw, L.-L.; Chiu, S.-H. Metschnikowia bicuspidata dominates in Taiwanese cold-weather yeast infections of Macrobrachium rosenbergii. Dis. Aquat. Org. 2007, 75, 191–199. [Google Scholar] [CrossRef]
- Wang, L.; Yue, L.; Chi, Z.; Wang, X. Marine killer yeasts active against a yeast strain pathogenic to crab Portunus trituberculatus. Dis. Aquat. Org. 2008, 80, 211–218. [Google Scholar] [CrossRef]
- Moore, M.M.; Strom, M. Infection and mortality by the yeast Metschnikowia bicuspidata var. bicuspidata in chinook salmon fed live adult brine shrimp (Artemia franciscana). Aquaculture 2003, 220, 43–57. [Google Scholar] [CrossRef]
- Zhao, R.; Wenjun, S.; Libao, W.; Hui, L.; Zhijun, Y.; Runhao, H.; Xugan, W.; Hui, S.; Yi, Q.; Jie, C.; et al. A preliminary study on the “Zombie disease” of Exopalaemon carinicauda. J. Fish. China 2022, 47, 099414. [Google Scholar] [CrossRef]
- Cao, G.; Bao, J.; Feng, C.; Li, X.; Lang, Y.; Xing, Y.; Jiang, H. First report of Metschnikowia bicuspidata infection in Chinese grass shrimp (Palaemonetes sinensis) in China. Transbound. Emerg. Dis. 2022, 69, 3133–3141. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Jiang, H.; Shen, H.; Xing, Y.; Feng, C.; Li, X.; Chen, Q. First description of milky disease in the Chinese mitten crab Eriocheir sinensis caused by the yeast Metschnikowia bicuspidata. Aquaculture 2021, 532, 735984. [Google Scholar] [CrossRef]
- Sun, N.; Bao, J.; Liang, F.; Liu, F.; Jiang, H.; Li, X. Prevalence of ‘milky disease’caused by Metschnikowia bicuspidata in Eriocheir sinensis in Panjin city, China. Aquac. Res. 2022, 53, 1136–1140. [Google Scholar] [CrossRef]
- Zhang, H.-Q.; Chi, Z.; Liu, G.-L.; Zhang, M.; Hu, Z.; Chi, Z.-M. Metschnikowia bicuspidate associated with a milky disease in Eriocheir sinensis and its effectitve treatment by Massoia lactone. Microbiol. Res. 2021, 242, 126641. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Luo, Z.; Zhong, W.; Hao, J.; Wang, Y.; Zhang, Z.; Geng, X. Studies on pathogen and its pathogenesis causing milky disease of Chinese mitten crab Eriocheir sinensis. Period. Ocean. Univ. China 2021, 51, 23–32. [Google Scholar] [CrossRef]
- Meng, Q.; Hou, L.; Zhao, Y.; Huang, X.; Huang, Y.; Xia, S.; Gu, W.; Wang, W. iTRAQ-based proteomic study of the effects of Spiroplasma eriocheiris on Chinese mitten crab Eriocheir sinensis hemocytes. Fish. Shellfish. Immunol. 2014, 40, 182–189. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jang, J.W.; Kim, K.J.; Maeng, P.J. TCA cycle-independent acetate metabolism via the glyoxylate cycle in Saccharomyces cerevisiae. Yeast 2011, 28, 153–166. [Google Scholar] [CrossRef]
- Sibirny, A.A. Yeast peroxisomes: Structure, functions and biotechnological opportunities. FEMS Yeast Res. 2016, 16, fow038. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Du, Z.; Lu, S.; Wang, Z.; He, X. Regulation of Cat8 in energy metabolic balance and glucose tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2023, 107, 4605–4619. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Church, M.; Hokamp, K.; Alhussain, M.M.; Bamagoos, A.A.; Fleming, A.B. Systematic analysis of tup1 and cyc8 mutants reveals distinct roles for TUP1 and CYC8 and offers new insight into the regulation of gene transcription by the yeast Tup1-Cyc8 complex. PLoS Genet. 2023, 19, e1010876. [Google Scholar] [CrossRef]
- Braam, S.; Tripodi, F.; Österberg, L.; Persson, S.; Welkenhuysen, N.; Coccetti, P.; Cvijovic, M. Exploring carbon source related localization and phosphorylation in the Snf1/Mig1 network using population and single cell-based approaches. Microb. Cell 2024, 11, 143. [Google Scholar] [CrossRef]
- Pérez-Díaz, A.J.; Vázquez-Marín, B.; Vicente-Soler, J.; Prieto-Ruiz, F.; Soto, T.; Franco, A.; Cansado, J.; Madrid, M. cAMP-Protein kinase A and stress-activated MAP kinase signaling mediate transcriptional control of autophagy in fission yeast during glucose limitation or starvation. Autophagy 2023, 19, 1311–1331. [Google Scholar] [CrossRef]
- Pradhan, A.; Avelar, G.M.; Bain, J.M.; Childers, D.S.; Larcombe, D.E.; Netea, M.G.; Shekhova, E.; Munro, C.A.; Brown, G.D.; Erwig, L.P. Hypoxia promotes immune evasion by triggering β-glucan masking on the Candida albicans cell surface via mitochondrial and cAMP-protein kinase A signaling. mBio 2018, 9, e01318. [Google Scholar] [CrossRef]
- Sandai, D.; Yin, Z.; Selway, L.; Stead, D.; Walker, J.; Leach, M.D.; Bohovych, I.; Ene, I.V.; Kastora, S.; Budge, S. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 2012, 3, e00495-12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Peng, J.; Ren, K.; Yu, Y.; Li, D.; She, X.; Liu, W. Divergent mitochondrial responses and metabolic signal pathways secure the azole resistance in Crabtree-positive and negative Candida species. Microbiol. Spectr. 2024, 12, e04042-23. [Google Scholar] [CrossRef]
- Turcotte, B.; Liang, X.B.; Robert, F.; Soontorngun, N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2009, 10, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Grüning, N.-M.; Rinnerthaler, M.; Bluemlein, K.; Mülleder, M.; Wamelink, M.M.; Lehrach, H.; Jakobs, C.; Breitenbach, M.; Ralser, M. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 2011, 14, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Le Moullac, G.; Le Groumellec, M.; Ansquer, D.; Froissard, S.; Levy, P. Haematological and phenoloxidase activity changes in the shrimp Penaeus stylirostrisin relation with the moult cycle: Protection against vibriosis. Fish. Shellfish. Immunol. 1997, 7, 227–234. [Google Scholar] [CrossRef]
- Li, W.; Dun, B.; Wang, Z.; Qu, J. A Modified Method for the Efficient and Fast Extraction of Total RNA from Saccharomyces cerevisiae with Hot-Phenol. Biotechnol. Bull. 2012, 12, 163–166. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Postma, E.D.; Couwenberg, L.G.F.; van Roosmalen, R.N.; Geelhoed, J.; de Groot, P.A.; Daran-Lapujade, P. Top-Down, Knowledge-Based Genetic Reduction of Yeast Central Carbon Metabolism. mBio 2022, 13, e0297021. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Satheesh, S.V.; Raghavendra, M.L.; Sadhale, P.P. The key enzyme in galactose metabolism, UDP-galactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet. Biol. 2007, 44, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Gravelat, F.N.; Cerone, R.P.; Baptista, S.D.; Campoli, P.V.; Choe, S.I.; Kravtsov, I.; Vinogradov, E.; Creuzenet, C.; Liu, H.; et al. Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides. J. Biol. Chem. 2014, 289, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Cyert, M.S. Calcineurin signaling in Saccharomyces cerevisiae: How yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 2003, 311, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Raiymbek, G.; An, S.; Khurana, N.; Gopinath, S.; Larkin, A.; Biswas, S.; Trievel, R.C.; Cho, U.S.; Ragunathan, K. An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase. eLife 2020, 9, e53155. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, P.; Cai, M.; Wu, D.; Zhang, M.; Chen, M.; Zhao, Y. Effects of microplastics on the innate immunity and intestinal microflora of juvenile Eriocheir sinensis. Sci. Total Environ. 2019, 685, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Guerra, P.; Vuillemenot, L.-A.P.; van Oppen, Y.B.; Been, M.; Milias-Argeitis, A. TORC1 and PKA activity towards ribosome biogenesis oscillates in synchrony with the budding yeast cell cycle. J. Cell Sci. 2022, 135, jcs260378. [Google Scholar] [CrossRef]
- Goranov, A.I.; Gulati, A.; Dephoure, N.; Takahara, T.; Maeda, T.; Gygi, S.P.; Manalis, S.; Amon, A. Changes in cell morphology are coordinated with cell growth through the TORC1 pathway. Curr. Biol. 2013, 23, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.S.; Hancock, T.J.; Lumsdaine, S.W.; Kauffman, S.J.; Mangrum, M.M.; Phillips, E.K.; Sparer, T.E.; Reynolds, T.B. Activation of Cph1 causes ss (1,3)-glucan unmasking in Candida albicans and attenuates virulence in mice in a neutrophil-dependent manner. PLoS Pathog. 2021, 17, e1009839. [Google Scholar] [CrossRef] [PubMed]
- Ballou, E.R.; Avelar, G.M.; Childers, D.S.; Mackie, J.; Bain, J.M.; Wagener, J.; Kastora, S.L.; Panea, M.D.; Hardison, S.E.; Walker, L.A. Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2016, 2, 16238. [Google Scholar] [CrossRef] [PubMed]
- Visser, W.F.; Van Roermund, C.W.; Ijlst, L.; Waterham, H.R.; Wanders, R.J. Metabolite transport across the peroxisomal membrane. Biochem. J. 2007, 401, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Bojunga, N.; Kötter, P.; Entian, K.-D. The succinate/fumarate transporter Acr1p of Saccharomyces cerevisiae is part of the gluconeogenic pathway and its expression is regulated by Cat8p. Mol. Gen. Genet. 1998, 260, 453–461. [Google Scholar] [CrossRef]
- Moosavi, B.; Zhu, X.-l.; Yang, W.-C.; Yang, G.-F. Molecular pathogenesis of tumorigenesis caused by succinate dehydrogenase defect. Eur. J. Cell Biol. 2020, 99, 151057. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Encarnacion-Rosado, J.; Kimmelman, A.C. Autophagy fuels mitochondrial function through regulation of iron metabolism in pancreatic cancer. Autophagy 2024, 20, 963–964. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Wang, Q.; Yang, F.; Liu, R.; Gao, Q.; Cheng, B.; Lin, X.; Huang, L.; Chen, C.; Xiang, J. Signaling metabolite succinylacetone activates HIF-1α and promotes angiogenesis in GSTZ1-deficient hepatocellular carcinoma. JCI Insight 2023, 8, e164968. [Google Scholar] [CrossRef] [PubMed]
- Bayar, I.; Asici, G.S.E.; Bildik, A.; Kiral, F. Gene Expression of Glycolysis Enzymes in MCF-7 Breast Cancer Cells Exposed to Warburg Effect and Hypoxia. Int. J. Mol. Cell. Med. 2024, 13, 29. [Google Scholar] [CrossRef]
- Bidault, G.; Virtue, S.; Petkevicius, K.; Jolin, H.E.; Dugourd, A.; Guénantin, A.-C.; Leggat, J.; Mahler-Araujo, B.; Lam, B.Y.; Ma, M.K. SREBP1-induced fatty acid synthesis depletes macrophages antioxidant defences to promote their alternative activation. Nat. Metab. 2021, 3, 1150–1162. [Google Scholar] [CrossRef]
- Shi, S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio 2014, 5, e01130. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ortiz, C.; Carrillo-Garmendia, A.; Correa-Romero, B.F.; Canizal-García, M.; González-Hernández, J.C.; Regalado-Gonzalez, C.; Olivares-Marin, I.K.; Madrigal-Perez, L.A. SNF1 controls the glycolytic flux and mitochondrial respiration. Yeast 2019, 36, 487–494. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Log2FC | FDR | Pfam Annotation | Swiss-Prot Annotation |
|---|---|---|---|---|
| METBIDRAFT_43176 | 13.96 | 4.54 × 10−20 | Calcineurin-like phosphoesterase | Putative metallophosphoesterase |
| METBIDRAFT_60496 | 4.79 | 3.70 × 10−6 | Cellulase (glycosyl hydrolase family 5) | Beta-xylosidase |
| METBIDRAFT_37387 | 3.85 | 0.000142613 | GPR1/FUN34/yaaH family | Glyoxylate pathway regulator |
| METBIDRAFT_227659 | 3.73 | 0.000292482 | Sugar (and other) transporter | Carboxylic acid transporter protein homolog |
| METBIDRAFT_33398 | 3.59 | 3.59 × 10−6 | Sugar (and other) transporter | Low-affinity glucose transporter HXT3 |
| METBIDRAFT_12194 | 3.26 | 0.000742583 | Sugar (and other) transporter | Sugar transporter STL1 |
| METBIDRAFT_29250 | 3.25 | 0.000763428 | Redoxin | Putative peroxiredoxin (Fragment) |
| METBIDRAFT_46728 | 3.22 | 0.000170095 | Mitochondrial carrier protein | Succinate/fumarate mitochondrial transporter |
| METBIDRAFT_76426 | 3.19 | 0.001763201 | Choline/Carnitine o-acyltransferase | Putative mitochondrial carnitine O-acetyltransferase |
| METBIDRAFT_70890 | 3.07 | 4.77 × 10−7 | Amino acid permease | General amino-acid permease GAP2 |
| METBIDRAFT_76181 | 2.98 | 4.20 × 10−17 | Major Facilitator Superfamily | Multidrug resistance protein 1 |
| METBIDRAFT_76577 | 2.92 | 0.000122001 | Sugar (and other) transporter | High-affinity glucose transporter HXT2 |
| METBIDRAFT_79721 | 2.80 | 0.000389761 | Sugar (and other) transporter | Major facilitator-type transporter ecdD |
| METBIDRAFT_79129 | 2.78 | 0.000217936 | GPR1/FUN34/yaaH family | Accumulation of dyads protein 2 |
| METBIDRAFT_33390 | 2.73 | 0.000286938 | Zinc finger, C2H2 type | Transcriptional regulator of yeast form adherence 4 |
| METBIDRAFT_30436 | 2.72 | 8.88 × 10−7 | Phosphate transporter family | Phosphate permease PHO89 |
| METBIDRAFT_40756 | 2.61 | 0.008238589 | Alpha amylase, catalytic domain | Alpha-glucosidase |
| METBIDRAFT_179260 | 2.54 | 2.15 × 10−8 | NA a | NA |
| METBIDRAFT_75879 | 2.53 | 0.000199658 | Amino acid permease | Proline-specific permease |
| METBIDRAFT_37464 | 2.51 | 1.44 × 10−7 | Mitochondrial import receptor subunit or translocase | Mitochondrial import receptor subunit TOM5 |
| Gene Name | Log2FC | FDR | Pfam Annotation | Swiss-Prot Annotation |
|---|---|---|---|---|
| METBIDRAFT_34094 | −13.78 | 8.25 × 10−29 | Cupin-like domain | JmjC domain-containing protein 4 |
| METBIDRAFT_78276 | −2.61 | 1.38 × 10−13 | Major Facilitator Superfamily | MFS antiporter QDR3 |
| METBIDRAFT_103598 | −2.61 | 0.00074696 | NA a | NA |
| METBIDRAFT_13862 | −2.49 | 0.000396594 | NA | NA |
| METBIDRAFT_137696 | −2.44 | 0.001872645 | NA | NA |
| METBIDRAFT_165330 | −2.37 | 1.51 × 10−15 | NA | NA |
| METBIDRAFT_169905 | −2.29 | 6.12 × 10−18 | NA | NA |
| METBIDRAFT_36205 | −2.23 | 1.30 × 10−29 | GATA zinc finger | Suppressor of ferric uptake 1 |
| METBIDRAFT_114380 | −2.12 | 2.10 × 10−21 | NA | NA |
| METBIDRAFT_36206 | −2.11 | 1.89 × 10−20 | NA | NA |
| METBIDRAFT_18009 | −2.08 | 1.51 × 10−10 | DDE superfamily endonuclease | Protein PDC2 |
| METBIDRAFT_46171 | −2.06 | 2.51 × 10−49 | Carbohydrate/starch-binding module (family 21) | NA |
| METBIDRAFT_153484 | −2.04 | 1.44 × 10−18 | NA | NA |
| METBIDRAFT_188454 | −2.02 | 3.01 × 10−14 | NA | NA |
| METBIDRAFT_110843 | −2.02 | 6.28 × 10−11 | NA | NA |
| METBIDRAFT_30879 | −2.00 | 3.01 × 10−26 | NA | NA |
| METBIDRAFT_10301 | −1.98 | 5.37 × 10−19 | NA | NA |
| METBIDRAFT_117096 | −1.97 | 1.59 × 10−22 | NA | NA |
| METBIDRAFT_10902 | −1.95 | 4.20 × 10−17 | Isocitrate lyase family | Isocitrate lyase |
| METBIDRAFT_31334 | −1.91 | 1.56 × 10−19 | Cytochrome P450 | Cytochrome P450 monooxygenase |
| Gene Symbol | geNorm | NormFinder | BestKeeper | Delta Ct | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S a | R a | S | R | S | R | S | R | S | R | |
| 18S | 1.284 | 4 | 0.654 | 4 | 0.858 | 1 | 1.284 | 4 | 2.99 | 4 |
| ACT1 | 1.068 | 2 | 0.381 | 2 | 1.075 | 2 | 1.068 | 2 | 2.21 | 2 |
| GAPDH | 1.307 | 5 | 0.730 | 6 | 1.290 | 3 | 1.307 | 5 | 3.08 | 5 |
| RIP | 1.336 | 6 | 0.669 | 5 | 1.985 | 6 | 1.336 | 6 | 5.73 | 6 |
| LSC2 | 1.191 | 3 | 0.552 | 3 | 1.548 | 5 | 1.191 | 3 | 2.59 | 3 |
| PMA1 | 1.059 | 1 | 0.263 | 1 | 1.467 | 4 | 1.059 | 1 | 2.00 | 1 |
| TAF10 | 1.704 | 7 | 1.068 | 7 | 2.124 | 7 | 1.704 | 7 | 7.00 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, B.; Zuo, B.; Li, X. Potential Strategies Applied by Metschnikowia bicuspidata to Survive the Immunity of Its Crustacean Hosts. Pathogens 2025, 14, 95. https://doi.org/10.3390/pathogens14010095
Zhang J, Li B, Zuo B, Li X. Potential Strategies Applied by Metschnikowia bicuspidata to Survive the Immunity of Its Crustacean Hosts. Pathogens. 2025; 14(1):95. https://doi.org/10.3390/pathogens14010095
Chicago/Turabian StyleZhang, Ji, Bingyu Li, Bingnan Zuo, and Xiaodong Li. 2025. "Potential Strategies Applied by Metschnikowia bicuspidata to Survive the Immunity of Its Crustacean Hosts" Pathogens 14, no. 1: 95. https://doi.org/10.3390/pathogens14010095
APA StyleZhang, J., Li, B., Zuo, B., & Li, X. (2025). Potential Strategies Applied by Metschnikowia bicuspidata to Survive the Immunity of Its Crustacean Hosts. Pathogens, 14(1), 95. https://doi.org/10.3390/pathogens14010095







