Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Real-Time PCR
2.3. Whole Genome Sequencing
2.4. Phylogenetic Analysis
3. Results
3.1. Case Diagnosis
3.2. Detection of Mpox Virus
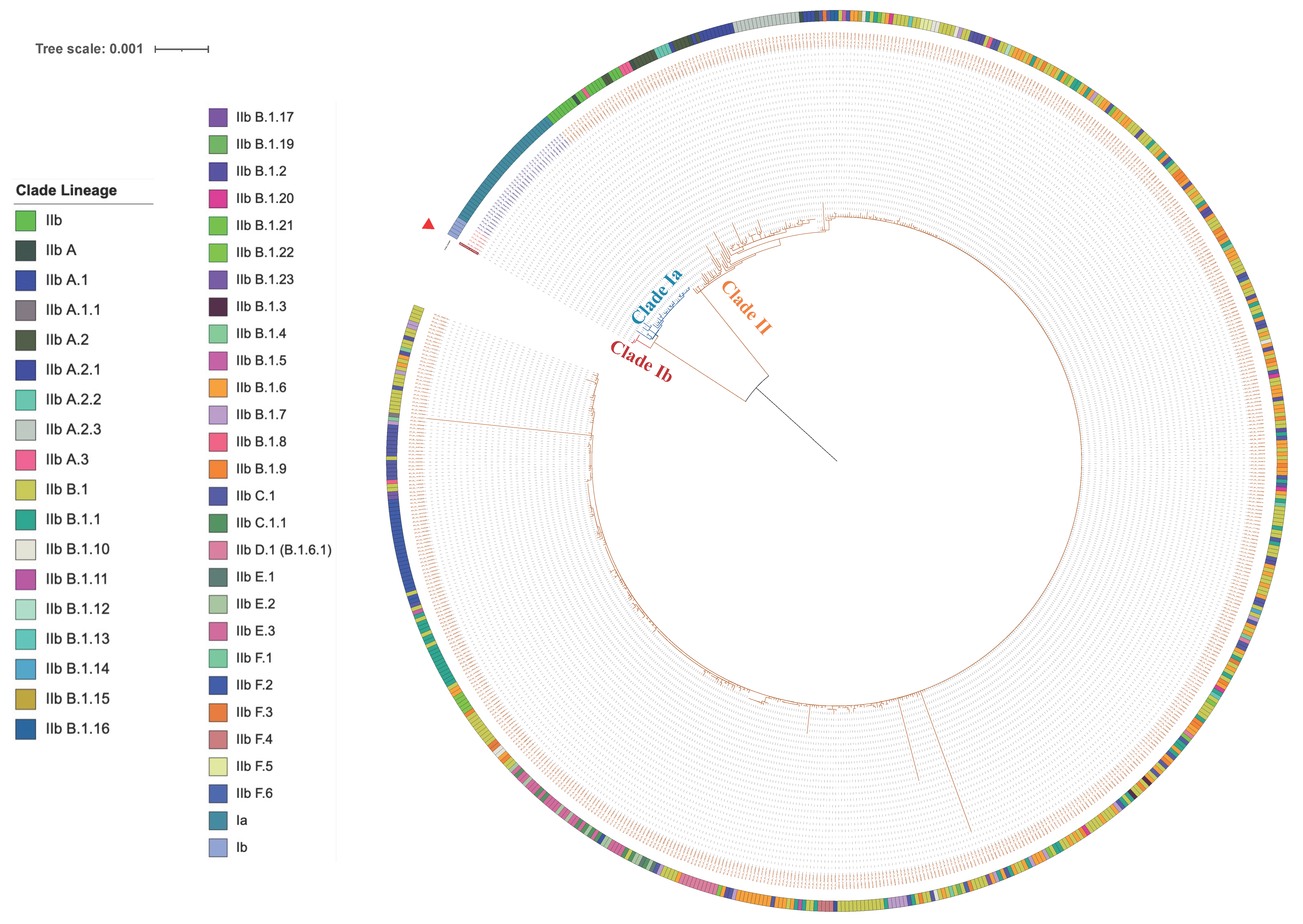
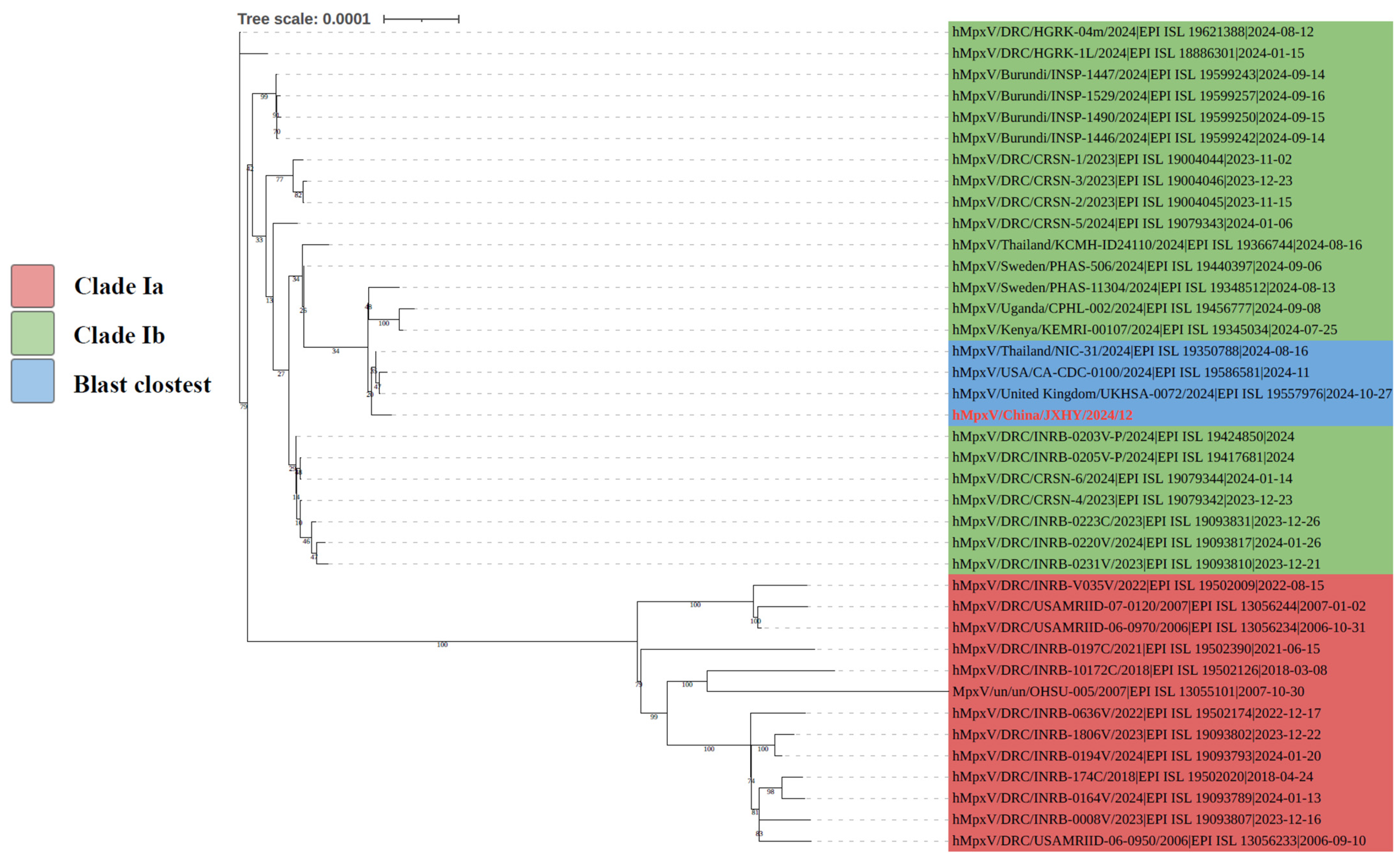
3.3. Analysis of Phylogenetic Tree
3.4. Analysis of Mutation Sites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- McCollum, A.M.; Damon, I.K. Human monkeypox. Clin. Infect. Dis. 2014, 58, 260–267. [Google Scholar] [CrossRef]
- Otu, A.; Ebenso, B.; Walley, J.; Barcelo, J.M.; Ochu, C.L. Global human monkeypox outbreak: Atypical presentation demanding urgent public health action. Lancet Microbe 2022, 3, e554–e555. [Google Scholar] [CrossRef]
- Cabanillas, B.; Murdaca, G.; Guemari, A.; Torres, M.J.; Azkur, A.K.; Aksoy, E.; Vitte, J.; de Las Vecillas, L.; Giovannini, M.; Fernandez-Santamaria, R.; et al. A compilation answering 50 questions on monkeypox virus and the current monkeypox outbreak. Allergy 2023, 78, 639–662. [Google Scholar] [CrossRef]
- Carrubba, S.; Geevarghese, A.; Solli, E.; Guttha, S.; Sims, J.; Sperber, L.; Meehan, S.; Ostrovsky, A. Novel severe oculocutaneous manifestations of human monkeypox virus infection and their historical analogues. Lancet Infect. Dis. 2023, 23, e190–e197. [Google Scholar] [CrossRef]
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar]
- Khodakevich, L.; Jezek, Z.; Messinger, D. Monkeypox virus: Ecology and public health significance. Bull. World Health Organ. 1988, 66, 747–752. [Google Scholar]
- Khodakevich, L.; Szczeniowski, M.; Manbuma, D.; Jezek, Z.; Marennikova, S.; Nakano, J.; Messinger, D. The role of squirrels in sustaining monkeypox virus transmission. Trop. Geogr. Med. 1987, 39, 115–122. [Google Scholar]
- Doty, J.B.; Malekani, J.M.; Kalemba, L.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Liyandja Dja Liyandja, T.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human-Animal Interface in the Democratic Republic of Congo. Viruses 2017, 9, 283. [Google Scholar] [CrossRef]
- Alakunle, E.F.; Okeke, M.I. Monkeypox virus: A neglected zoonotic pathogen spreads globally. Nat. Rev. Microbiol. 2022, 20, 507–508. [Google Scholar] [CrossRef]
- Nuzzo, J.B.; Borio, L.L.; Gostin, L.O. The WHO Declaration of Monkeypox as a Global Public Health Emergency. JAMA 2022, 328, 615–617. [Google Scholar] [CrossRef]
- Sabeena, S. The changing epidemiology of monkeypox and preventive measures: An update. Arch. Virol. 2023, 168, 31. [Google Scholar] [CrossRef]
- Papukashvili, D.; Rcheulishvili, N.; Liu, C.; Wang, X.; He, Y.; Wang, P.G. Strategy of developing nucleic acid-based universal monkeypox vaccine candidates. Front. Immunol. 2022, 13, 1050309. [Google Scholar] [CrossRef]
- Li, H.; Zhang, H.; Ding, K.; Wang, X.H.; Sun, G.Y.; Liu, Z.X.; Luo, Y. The evolving epidemiology of monkeypox virus. Cytokine Growth Factor Rev. 2022, 68, 1–12. [Google Scholar] [CrossRef]
- Condit, R.C.; Moussatche, N.; Traktman, P. In a nutshell: Structure and assembly of the vaccinia virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the monkeypox virus genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, Y.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A tale of two clades: Monkeypox viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef]
- Andrei, G.; Snoeck, R. Differences in pathogenicity among the mpox virus clades: Impact on drug discovery and vaccine development. Trends Pharmacol. Sci. 2023, 44, 719–739. [Google Scholar] [CrossRef]
- Haller, S.L.; Peng, C.; McFadden, G.; Rothenburg, S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2014, 21, 15–40. [Google Scholar] [CrossRef]
- Locker, J.K.; Kuehn, A.; Schleich, S.; Rutter, G.; Hohenberg, H.; Wepf, R.; Griffiths, G. Entry of the two infectious forms of vaccinia virus at the plasma membane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 2000, 11, 2497–2511. [Google Scholar] [CrossRef]
- Bunge, E.M.; Hoet, B.; Chen, L.; Lienert, F.; Weidenthaler, H.; Baer, L.R.; Steffen, R. The changing epidemiology of human monkeypox-A potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022, 16, e0010141. [Google Scholar] [CrossRef]
- Meo, S.A.; Al-Khlaiwi, T.; Al Jassir, F.F.; Meo, A.S. Impact of traveling on transmission trends of human monkeypox disease: Worldwide data based observational analysis. Front. Public Health 2023, 11, 1029215. [Google Scholar] [CrossRef] [PubMed]
- Costello, V.; Sowash, M.; Gaur, A.; Cardis, M.; Pasieka, H.; Wortmann, G.; Ramdeen, S. Imported Monkeypox from International Traveler, Maryland, USA, 2021. Emerg. Infect. Dis. 2022, 28, 1002–1005. [Google Scholar] [CrossRef] [PubMed]
- Adetifa, I.; Muyembe, J.J.; Bausch, D.G.; Heymann, D.L. Mpox neglect and the smallpox niche: A problem for Africa, a problem for the world. Lancet 2023, 401, 1822–1824. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Acharya, A.; Gendelman, H.E.; Byrareddy, S.N. The 2022 outbreak and the pathobiology of the monkeypox virus. J. Autoimmun. 2022, 131, 102855. [Google Scholar] [CrossRef]
- Alakunle, E.; Moens, U.; Nchinda, G.; Okeke, M.I. Monkeypox Virus in Nigeria: Infection Biology, Epidemiology, and Evolution. Viruses 2020, 12, 1257. [Google Scholar] [CrossRef]
- Martinez-Fernandez, D.E.; Fernandez-Quezada, D.; Casillas-Munoz, F.A.G.; Carrillo-Ballesteros, F.J.; Ortega-Prieto, A.M.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A. Human Monkeypox: A Comprehensive Overview of Epidemiology, Pathogenesis, Diagnosis, Treatment, and Prevention Strategies. Pathogens 2023, 12, 947. [Google Scholar] [CrossRef]
- Malik, S.; Ahmed, A.; Ahsan, O.; Muhammad, K.; Waheed, Y. Monkeypox Virus: A Comprehensive Overview of Viral Pathology, Immune Response, and Antiviral Strategies. Vaccines 2023, 11, 1345. [Google Scholar] [CrossRef]
- Zahmatyar, M.; Fazlollahi, A.; Motamedi, A.; Zolfi, M.; Seyedi, F.; Nejadghaderi, S.A.; Sullman, M.J.M.; Mohammadinasab, R.; Kolahi, A.A.; Arshi, S.; et al. Human monkeypox: History, presentations, transmission, epidemiology, diagnosis, treatment, and prevention. Front. Med. 2023, 10, 1157670. [Google Scholar] [CrossRef]
- Dou, X.; Li, F.; Ren, Z.; Zhang, D.; Li, J.; Li, D.; Sun, Y.; Jin, H.; Li, R.; Li, W.; et al. Clinical, epidemiological, and virological features of Mpox in Beijing, China—31 May–21 June 2023. Emerg. Microbes Infect. 2023, 12, 2254407. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, X.; Liu, J.; Xiang, L.; Huang, S.; Xie, X.; Fang, L.; Lin, Y.; Zhang, M.; Wang, L.; et al. Phylogeny and molecular evolution of the first local monkeypox virus cluster in Guangdong Province, China. Nat. Commun. 2023, 14, 8241. [Google Scholar] [CrossRef]
- Nitiyanontakij, R.; Reawrang, S.; Kaewsawang, P.; Suwanwattana, P.; Prasithsirikul, W.; Pongpirul, W.; Sunchatawirul, K. Monkeypox Virus Isolate Monkeypox Virus/Human/Thailand/DDCBIDI-002/2024, Partial Genome. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/PQ281491.1?report=genbank&log$=nucltop&blast_rank=16&RID=RCF63DGA016 (accessed on 4 September 2024).
- Chen, N.; Li, G.; Liszewski, M.K.; Atkinson, J.P.; Jahrling, P.B.; Feng, Z.; Schriewer, J.; Buck, C.; Wang, C.; Lefkowitz, E.J.; et al. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 2005, 340, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Hutson, C.L.; Abel, J.A.; Carroll, D.S.; Olson, V.A.; Braden, Z.H.; Hughes, C.M.; Dillon, M.; Hopkins, C.; Karem, K.L.; Damon, I.K.; et al. Comparison of West African and Congo Basin monkeypox viruses in BALB/c and C57BL/6 mice. PLoS ONE 2010, 5, e8912. [Google Scholar] [CrossRef] [PubMed]
- Sah, R.; Abdelaal, A.; Reda, A.; Katamesh, B.E.; Manirambona, E.; Abdelmonem, H.; Mehta, R.; Rabaan, A.A.; Alhumaid, S.; Alfouzan, W.A.; et al. Monkeypox and Its Possible Sexual Transmission: Where Are We Now with Its Evidence? Pathogens 2022, 11, 924. [Google Scholar] [CrossRef]
- Hammarlund, E.; Dasgupta, A.; Pinilla, C.; Norori, P.; Fruh, K.; Slifka, M.K. Monkeypox virus evades antiviral CD4+ and CD8+ T cell responses by suppressing cognate T cell activation. Proc. Natl. Acad. Sci. USA 2008, 105, 14567–14572. [Google Scholar] [CrossRef]
- Durski, K.N.; McCollum, A.M.; Nakazawa, Y.; Petersen, B.W.; Reynolds, M.G.; Briand, S.; Djingarey, M.H.; Olson, V.; Damon, I.K.; Khalakdina, A. Emergence of Monkeypox—West and Central Africa, 1970–2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 306–310. [Google Scholar] [CrossRef]
- Wang, Y.; Leng, P.; Zhou, H. Global transmission of monkeypox virus-a potential threat under the COVID-19 pandemic. Front. Immunol. 2023, 14, 1174223. [Google Scholar] [CrossRef]
- Kumar, P.; Chaudhary, B.; Yadav, N.; Devi, S.; Pareek, A.; Alla, S.; Kajal, F.; Nowrouzi-Kia, B.; Chattu, V.K.; Gupta, M.M. Recent Advances in Research and Management of Human Monkeypox Virus: An Emerging Global Health Threat. Viruses 2023, 15, 937. [Google Scholar] [CrossRef]
- World Health Organization. Multi-Country Monkeypox Outbreak in Non-Endemic Countries. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385 (accessed on 21 May 2022).
| Specimen | Source | CT |
|---|---|---|
| Herpes swab | Face | 26.09 |
| Oropharyngeal swab | Oropharynx | 26.50 |
| Herpes swab | Protothorax | 27.94 |
| Herpes swab | Abdomen | 26.78 |
| Herpes swab | Back | 18.63 |
| Herpes swab | Left arm | 19.04 |
| Herpes swab | Left buttock | 18.10 |
| Herpes swab | Left lower extremity | 20.08 |
| Herpes swab | Right arm | 18.11 |
| Herpes swab | Right femoral | 19.59 |
| Herpes swab | Right lower extremity | 19.03 |
| Plasma | Blood | 34.08 |
| Full blood | Blood | Negative |
| Position (bp) | Gene | Nucleotide Mutations | Amino Acid Mutations |
|---|---|---|---|
| 1368 | OPG001 | 257A>C | N86T |
| 3192 | OPG003 | 1460C>T | T487I |
| 5377 | OPG015 | 745G>A | A249T |
| 5979 | OPG015 | 143T>C | I48T |
| 6111 | OPG015 | 11T>C | V4A |
| 7900 | OPG019 | 334G>T | G112C |
| 11,487 | OPG023 | 1583G>A | C528Y |
| 16,320 | OPG027 | 237A>C | L79F |
| 21,582 | OPG034 | 551C>A | P184H |
| 23,143 | OPG036 | 49G>A | V17I |
| 27,161 | OPG040 | 480G>A | M160I |
| 28,787 | OPG042 | 476C>T | T159I |
| 29,918 | OPG043 | 203A>G | N68S |
| 36,113 | OPG052 | 29C>G | T10S |
| 36,669 | OPG053 | 168G>A | M56I |
| 38,108 | OPG054 | 35G>A | R12Q |
| 48,904 | OPG066 | 664C>A | L222I |
| 49,148 | OPG066 | 420G>T | K140N |
| 53,006 | OPG070 | 34T>C | Y12H |
| 73,259 | OPG089 | 1009A>G | K337E |
| 78,198 | OPG096 | 224T>C | L75P |
| 80,796 | OPG100 | 401G>A | R134H |
| 97,180 | OPG115 | 124T>C | Y42H |
| 99,175 | OPG117 | 730G>A | E244K |
| 103,775 | OPG120 | 371C>T | S124L |
| 108,574 | OPG125 | 1478C>T | T493M |
| 110,453 | OPG126 | 76G>T | A26S |
| 110,647 | OPG127 | 577T>C | Y193H |
| 119,766 | OPG136 | 2220G>A | M740I |
| 124,029 | OPG140 | 116C>A | P39H |
| 126,571 | OPG145 | 227C>A | P76Q |
| 130,270 | OPG150 | 94G>A | D32N |
| 132,139 | OPG151 | 818G>A | R273Q |
| 140,136 | OPG155 | 302T>G | I101R |
| 140,573 | OPG156 | 783G>A | M261I |
| 145,111 | OPG164 | 392A>T | N131I |
| 145,155 | OPG164 | 436G>A | V146I |
| 148,457 | OPG170 | 286G>A | V96I |
| 150,078 | OPG173 | 62A>G | N21S |
| 150,101 | OPG173 | 85A>G | I29V |
| 150,383 | OPG174 | 994G>A | D332N |
| 150,434 | OPG174 | 943C>T | R315C |
| 159,350 | OPG185 | 716C>T | T239I |
| 161,996 | OPG188 | 680C>T | S227F |
| 165,883 | OPG191 | 3A>G | I1M |
| 171,226 | OPG199 | 426A>G | I142M |
| 172,116 | OPG200 | 155G>A | G52D |
| 179,159 | OPG208 | 101T>C | V34A |
| 182,565 | OPG210 | 1424C>T | A475V |
| 184,309 | OPG210 | 3168G>T | E1056D |
| 185,991 | OPG210 | 4850G>A | G1617E |
| 186,727 | OPG210 | 5586T>G | D1862E |
| 190,857 | OPG015_dup | 11T>C | V4A |
| 190,989 | OPG015_dup | 143T>C | I48T |
| 191,591 | OPG015_dup | 745G>A | A249T |
| 193,776 | OPG003_dup | 1460C>T | T487I |
| 195,600 | OPG001_dup | 257A>C | N86T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Yan, Y.; Xu, J.; Lv, S.; Ren, G.; Zhou, Y.; Song, W.; Ge, R.; Xu, P.; Zhu, G.; et al. Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China. Pathogens 2025, 14, 102. https://doi.org/10.3390/pathogens14010102
Song Y, Yan Y, Xu J, Lv S, Ren G, Zhou Y, Song W, Ge R, Xu P, Zhu G, et al. Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China. Pathogens. 2025; 14(1):102. https://doi.org/10.3390/pathogens14010102
Chicago/Turabian StyleSong, Yin, Yong Yan, Jingyu Xu, Shencong Lv, Ganglin Ren, Yamei Zhou, Wanchen Song, Rui Ge, Peihua Xu, Guoying Zhu, and et al. 2025. "Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China" Pathogens 14, no. 1: 102. https://doi.org/10.3390/pathogens14010102
APA StyleSong, Y., Yan, Y., Xu, J., Lv, S., Ren, G., Zhou, Y., Song, W., Ge, R., Xu, P., Zhu, G., & Chen, Z. (2025). Complete Genome Sequence Analysis of the First Imported Mpox Virus Clade Ib Variant in China. Pathogens, 14(1), 102. https://doi.org/10.3390/pathogens14010102






