Abstract
The cluster of genes determining the production of botulinum toxins is an attribute of not only the Clostridium botulinum species. This cluster is also found in other members of the Clostridium genus, such as C. baratii, C. butyricum, and C. sporogenes. The occurrence of a botulinum-like cluster has also been recorded in strains of other genera, i.e., Enterococcus faecium, as well as in a Gram-negative species isolated from freshwater sediments; however, the biological activity of bont-related genes has not been noted. It can be said that the mentioned species have a dual nature. Another species with a dual nature is C. butyricum. This bacterium is a common human and animal gut commensal bacterium and is also frequently found in the environment. Although non-toxigenic strains are currently used as probiotics in Asia, other strains have been implicated in pathological conditions, such as botulism in infants or necrotizing enterocolitis in preterm neonates. Additionally, C. baratii strains are rare opportunistic pathogens associated with botulism intoxication. They have been isolated from food and soil and can be carried asymptomatically or cause botulism outbreaks in animals and humans. In addition to the mentioned clostridia, the other microorganisms considered as non-toxigenic have also been suspected of carrying botulinum cluster Gram-negative bacteria, such as Chryseobacterium piperi isolated from freshwater sediments; however, the biological activity of bont-related genes has not been noted. Additionally, Enterococcus faecium strains have been discovered carrying BoNT-related clusters (BoNT/En). Literature data regarding the heterogeneity of BoNT-producing strains indicate the requirement to reclassify C. botulinum species and other microorganisms able to produce BoNTs or possess botulinum-like gene clusters. This article aims to show the dual nature of Clostridium strains not belonging to the C. botulinum species that are sporadically able to carry bont clusters, which are usually considered saprophytic and even probiotic, and bont-like clusters in microorganisms from other genera. The aim was also to consider the genetic mechanisms of botulinum cluster expression in strains that are considered opportunistic and the microbiological safety aspects associated with their occurrence in the environment.
1. Introduction
Clostridium is a broad group of obligate anaerobes belonging to the Firmicutes bacterial phylum. Most of them have a Gram-positive cell wall structure. This genus includes mainly saprophytes. Some strains of this genus are considered probiotics but also significant human and animal pathogens that cause dangerous diseases, such as botulism, gangrene, and tetanus. They produce spores that are resistant to pasteurization temperatures. This genus inhabits the soil and the digestive tract of animals and humans. Clostridium is also found in the microflora of the female reproductive system. The genus Clostridium is a highly heterogeneous group of bacteria. Most of them are not associated with any pathogenic processes. Most species are saprophytes or even considered probiotics, even though some strains of this species could possess pathogenic features, such as an ability to produce botulinum toxins [1,2].
Historically, botulinum toxin was considered to be produced by Clostridium botulinum. Botulism has been affecting human civilization from the earliest time. However, an accurately described outbreak of foodborne botulism was reported as late as the 18th century [3]. The first isolation of C. botulinum was conducted in 1895 by Emile Piere van Ermengem. This pathogen was isolated from a salted ham associated with a botulism outbreak. Firstly, this microorganism was named Bacillus Botulinus and was changed further into C. botulinum [3]. The chronology of discovered toxinotypes of this bacterium (besides B and A) is indicated by subsequent letters of the alphabet from A to G. The bacterium discovered by van Ermengem was marked as B, and the last as G [4]. Based on these discoveries, a definition was formulated that all clostridia able to produce botulinum toxins are classified to C. botulinum species [5]. Due to differences in metabolism and 16S rRNA gene sequences, strains of this pathogen are classified into four metabolic groups, i.e., group I includes all type A and proteolytic type B and F strains, group II includes all type E and non-proteolytic type B and F strains, and group III consists of type C and D and mosaic strains—CD and DC. Group IV includes type G strains. The mentioned groups are related to other species considered to be non-toxigenic, i.e., C. sporogenes and C. tepidum are deemed related to group I, C. butyricum, C. taeniosporium, and C. beijerinckii to group II, C. novyi to group III, and C. argentinense, C. subterminale, and C. schirmacherense to group IV [6,7].
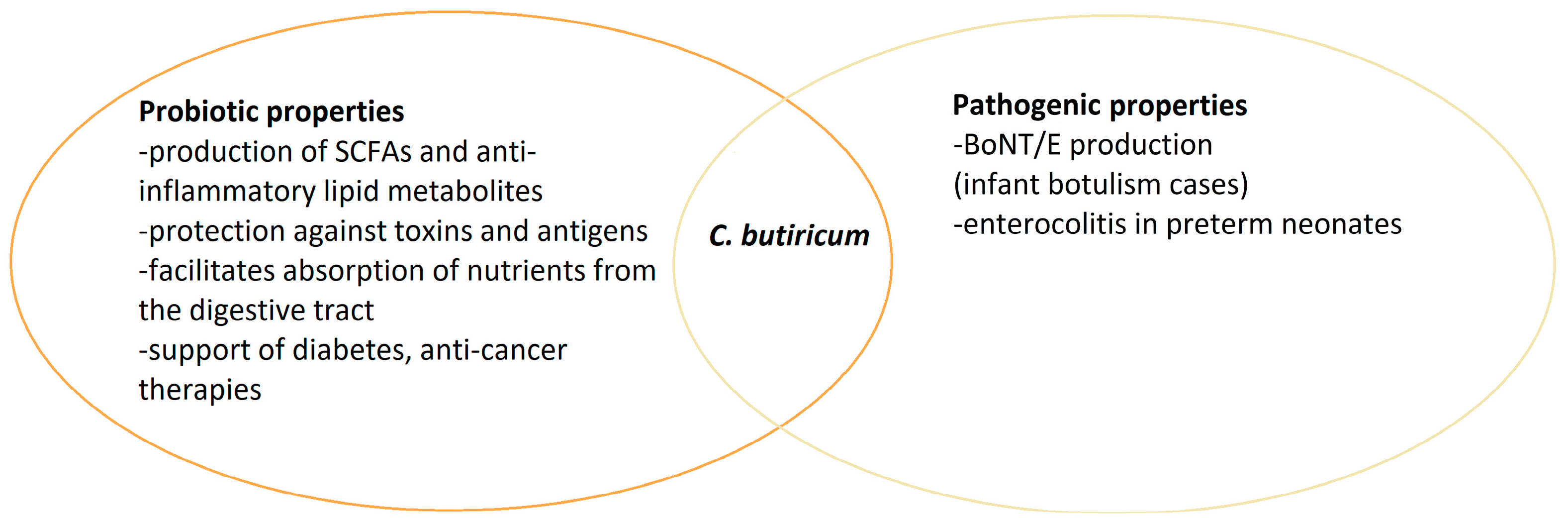
Moreover, it has been observed that horizontal transfer of genes determining toxigenicity between strains of C. botulinum and related species is possible (Figure 1). The ability to produce botulinum toxins has been previously reported in some strains of C. butyricum and C. baratii [7]. Some strains of C. butyricum can produce BoNT/E. The evidence of toxigenic C. butyricum isolation has been noted in the cases of intestinal toxemia botulism in infants and adults in Italy, Japan, the USA, Ireland, and Great Britain [8]. In recent years, it has also been reported in C. sporogenes strains [9]. The detection of C. botulinum is not possible solely based on biochemical features. It is difficult due to the diversity mentioned above within the genus, i.e., the occurrence of strains phenotypically similar to this species, which cannot produce BoNTs. It should also be noted that the first thing generally determined about a BoNT-producing organism is that it produces a toxin, followed by its toxin type, and then the organism genus and species. Moreover, the toxigenicity of group III strains (toxinotypes C, D and their mosaic CD and DC variants) is determined by the conversion of lysogenic bacteriophages, and the production of toxins by the group I and II strains may be conditioned by horizontal gene transfer [6]. Defining C. botulinum as a species is challenging because of the mentioned heterogeneity and phenotypical diversity.
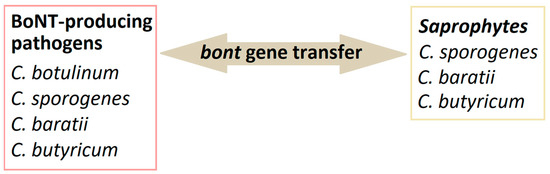
Figure 1.
Horizontal bont gene transfer among Clostridium strains.
2. Saprophytic and Pathogenic Clostridium sporogenes
2.1. General Clostridium sporogenes Description
Clostridium sporogenes is an anaerobic Gram-positive, spore-producing rod found in soil and the human and animal gastrointestinal tract as a part of the normal intestinal flora [10] and is closely related to proteolytic strains of Clostridium botulinum. The spoilage of this organism usually results in blown or torn packages with a strong, putrid odor, first described in detail by Metchnikoff in the year 1908. The first strains of C. sporogenes have been isolated from the gastrointestinal tract of healthy individuals and those with chronic colitis [11].
C. sporogenes species include both pathogenic and saprophytic strains. C. sporogenes cause food spoilage due to their genetic and physiological similarity to Clostridium botulinum group I. Some strains of both C. sporogenes and C. botulinum group I produce spores that are highly resistant to heat in the environment. This state of metabolic dormancy combined with heat resistance allows these bacteria to survive in adverse conditions such as lack of nutrients, desiccation, oxygen, high pressure, heat treatment, and toxic chemicals. It is why these bacteria pose severe problems with food spoilage and food safety [12].
2.2. Pathogenic Strains of Clostridium sporogenes
Pathogenic activity of C. sporogenes strains is most often described in immunocompromised patients, e.g., HIV-positive or cancer patients, COVID patients with pneumonia, or elderly (older than 65 years) people [10,13,14,15,16]. C. sporogenes infections have also been detected in patients with leukopenia and renal transplant recipients [12]. The most severe cases associated with pathogenic strains of C. sporogenes are related to bacteremia. According to our knowledge, up to now, there have been 29 described infections caused by C. sporogenes, including 19 bacteremias, one pyogenic liver abscess, two empyemas, one septic arthritis, two septicaemias [17], and four gas gangrenes [12,14,15,17,18,19,20,21,22,23,24,25]. In the case of bacteremia, most cases have occurred in immunocompromised patients (17 cases) [14,18,19]. Bacterial mortality is most likely related to the production of a hemorrhagic toxin and proteinases. The C. sporogenes hemorrhagic toxin is maximally produced at the early growth phase, is produced well in a peptone- or ammonia-rich medium, and has a molecular weight different from that of other intestinal hemorrhage-inducing bacterial toxins released by C. difficile, C. perfringens, and C. sordellii [26]. Hara-Kudo et al. studies have shown that the hemorrhagic toxin has collagenase activity and is responsible for the hydrolysis of type III and collagen IV, the main component of blood vessels’ intima and tunica media. Therefore, the hemorrhagic toxin produced by C. sporogenes is an important virulence factor, and the hemorrhage induced by this toxin is related to its collagenase activity [26]. On the other hand, identifying C. sporogenes as a cause of bacteremia in an immunocompetent patient is very rare. In immunocompetent patients, a C. sporogenes infection has resulted from a secondary infection [12,15].
Clostridium sporogenes and C. botulinum Group I are closely related mesophilic bacteria with genotypic and physiological characteristics, including proteolytic properties and the ability to form spores of high thermal resistance. Clostridium botulinum Group I and C. sporogenes are responsible for foodborne, infant, and wound botulism [27]. Similar to C. botulinum group I and II strains, some C. sporogenes strains could produce botulinum neurotoxin serotype B. The genes encoding the production of all BoNT/B subtypes are organized in a haemagglutinin gene cluster. However, depending on the subtype present, it may be located within the chromosome or carried by mobile extrachromosomal elements [28]. Research by previous authors has shown that C. sporogenes strains carrying the bont/B1-B6 genes occurred in different geographical regions, e.g., France, Australia, the United Kingdom, Italy, and the United States [28,29,30,31,32,33]. Moreover, strains responsible for infant infections (strain no. CDC 1632 and AMA1195) [28,33] and infection of wounds (stain no. B2 450) have also been mentioned [32]. Brunt et al. have reported that the plasmid-borne subtypes BoNT/B1, B2, and B6 could be produced by C. botulinum and C. sporogenes. A comparative genomic study with 556 highly diverse strains of C. botulinum Group I and C. sporogenes (including 417 newly sequenced strains) has reported a core genome single-nucleotide polymorphism (SNP) analysis that revealed two significant lineages: C. botulinum Group I (most strains possessed botulinum neurotoxin gene(s) of types A, B, and F) and C. sporogenes (some strains possessed a type B botulinum neurotoxin gene). Of the 104 C. sporogenes strains identified, 20 isolates possessed a gene encoded botulinum neurotoxin or either subtype B1, B2, or B6. All subtype B1, B2, and B6 bont genes were in a ha neurotoxin gene cluster. Moreover, a new cluster containing the gene encoding the botulinum toxin subtype B1 has been identified in five C. sporogenes strains [27].
3. Double Nature of Clostridium butyricum
Clostridium butyricum is an obligate, anaerobic, rod-shaped Gram-positive bacterium, spore-forming in various environments, such as the human gastrointestinal tract and soil. These organisms have many applications in fuel industries as by-product producers and in medicine as probiotic strains, especially in Asia [34]. Butyric acid is the main chemical these bacteria produce in fermentation via the but–buk pathway of dietary fiber and other substances not digested by the human digestive system. Many studies indicate that short-chain fatty acids (SCAFs) produced in the intestines by the microbiota, which include butyric acid, influence the regulation of the homeostasis of the immune system and the functioning of the intestinal barrier [35]. Butyric acid is widely researched and known to have anti-inflammatory and anti-cancer properties. Table 1 showed the role of Clostridium butyricum in human health and disease. However, some strains of these organisms have also proved to be neurotoxigenic pathogens [8].
3.1. Clostridium butyricum as a Probiotic
Clostridium butyricum is commonly recommended as a probiotic after antibiotic therapy or surgical procedures. It was first isolated from pig intestines by Prażmowski in 1880 [36], and in 1933 it was isolated from the feces of healthy individuals by Dr Miyairi. Then, in 1963, C. butyricum MIYAIRI 588 (CBM 588) was isolated from the soil and found application as a probiotic strain. It was used in Japan as a drug to relieve gastrointestinal symptoms after antibiotic therapy, such as diarrhea. In 2014, the European Parliament authorized placing Clostridium butyricum (CBM 588) on the market as a new food ingredient under Regulation (EC) No 258/97 [37].
CBM 588 is entirely safe, which has been confirmed in laboratory tests on animals as a feed additive and in humans [38]. CBM 588 is resistant to stress effects such as low pH and antimicrobial agents [39]. After oral administration, the composition of the normal gastrointestinal microflora is regulated by increasing the beneficial microflora and reducing harmful strains of microorganisms. At the same time, it improves digestion and the functioning of the digestive system. They create an unfavorable environment for pathogenic organisms mainly by producing butyric acid and adhering to human epithelial cells, creating a protective mucosal barrier. It reduces the risk of infection development [39,40]. They can strengthen the intestinal immune response by stimulating the development of beneficial intestinal microflora [40]. A combination of probiotic strains can also be used to improve the effect of Clostridium as a probiotic. So et al. [41] investigated selecting specific strains of lactic acid bacteria (LAB) that could synergistically enhance the probiotic functions of C. butyricum. Supernatants of 249 lactic acid bacteria were examined, and observations were made that 24 strains did not inhibit the growth of C. butyricum. Additionally, 4 of these 24 strains induced a more than twofold promotion in the growth rate of C. butyricum during co-culture with this bacterial strain. This growth promotion was verified by qPCR. In particular, Lactobacillus brevis JL16 and Lactobacillus parabuchneri MH44 stimulated C. butyricum more effectively than other strains did [41].
CBM 588 is a probiotic bacterium that has already been used to prevent post-antibiotic diarrhea and in animal supplementation. Much research is still being conducted to elucidate the exact mechanism of protection of the intestinal epithelium by bacteria. CMB 588 is known to increase the abundance of Bifidobacterium, Lactobacillus, and Lactococcus and enhance intestinal barrier function in mice with dysbiosis after antibiotic therapy [40]. CMB 588 also significantly controls antibiotic-induced intestinal inflammation by increasing anti-inflammatory lipid metabolites such as palmitic acid, 15d-prostaglandin J2, and protectin D1 [40]. Studies also indicate that the administration of Clostridium butyricum effectively restores the intestinal microbial balance after colonoscopy and contributes to faster recovery [42].
Clostridium butyricum (CB) is also used as a supplement regulating the composition of the intestinal microflora and the quality of animal meat [43]. Zhang et al. investigated the effects of CB supplementation and rumen-protected fat (RPF) in increasing diet density and providing essential fatty acids on goat meat’s growth performance, nutritional value, and oxidative stability. The test results indicate that only in the case of shear force is there an observed interaction between CB and RPF. The content of intramuscular fat (IMF) is higher in the case of CB and RPF diets. Moreover, pH after 24 h and a* (redness) values, total antioxidant capacity, glutathione peroxidase activity, 18:3, 20:5, and total polyunsaturated fatty acid concentrations are increased, while L* (lightness) values, shear force, and malondialdehyde content (p = 0.044) are decreased by the addition of CB. In addition, CB supplementation increases the content of essential amino acids, flavor amino acids, and total amino acids. It also increases the expression of lipoprotein lipase and peroxisome proliferator-activated γ receptor (PPARγ) and decreases the expression of stearoyl-CoA desaturase (SCD). It should, therefore, be emphasized that the supplementation of CB and RPF in goats improves carcass characteristics, meat quality, and fat deposition in connection to increasing the expression of lipogenic genes in LT muscle [43]. Therefore, it is worth including these bacteria in the goat diet.
C. butyricum supplementation may also improve lipid metabolism and growth performance in piglets with intrauterine growth restriction (IUGR) and their suckling efficiency [44]. Zhang et al. investigated the effect of Clostridium supplementation on hepatic lipid disorders in IUGR-suckling piglets. The control sample consisted of piglets that received physiological saline added to milk. In contrast, the test sample consisted of piglets fed with milk with the addition of C. butyricum at a dose of 2 × 108 colony-forming units (CFU)/kg body weight. The research results indicate that C. butyricum supplementation influences the intestinal microflora of IUGR piglets, reducing opportunistic pathogens in the ileum, such as Streptococcus and Enterococcus. The microorganisms hydrolyze bile salts, increasing bile acids. These can be transported to the liver and act as signaling molecules to activate the hepatic X receptor α (LXRα) and farnesoid X receptor (FXR). Therefore, reducing the number of these microorganisms accelerates the synthesis and oxidation of fatty acids. It lowers cholesterol levels, which improves the morphological condition of the liver in IUGR piglets, normalizes lipid metabolism, and improves the suckling efficiency of IUGR piglets [44].
Studies also indicate the hepatoprotective effect of CB in sea bass by reducing the activity of hepatic aspartate aminotransferase (AST) and increasing alkaline reaction phosphatase (AKP) and acid phosphatase (ACP) activity. Additionally, CB has a significant impact on strengthening the livers’ immunity. CB regulates the content of metabolic biomarkers such as arachidonate, crotonyl-CoA, and D-glucose 1-phosphate, which affects the main gluconeogenic, lipogenic, and amino acid metabolic pathways [45]. Research results indicate improved immunity and metabolism in sea bass, suggesting a hepatoprotective effect in humans [45].
3.2. Role of Clostridium butyricum in Health Promotion
More and more research indicates that the intestinal microflora influences metabolic syndrome development. One of the pathologies of this disease is obesity. Research suggests a negative correlation between the content of C. butyricum and the predisposition to the development of obesity, but the mechanism of this effect is still unclear. Five isolates of C. butyricum were administered (FYNDL1T1 (L1T1) and FHBSJZ1T1 (Z1T1) were isolated from the feces of cow and dog, respectively, strains FHLJZD47T7 (47T7) and NXYCHL3M3 (L3M3) were isolated from healthy volunteers, and strain C20_1_1 (C20) was isolated from an obese volunteer) to mice on a high-fat diet to determine their impact on the development of obesity. All tested isolates inhibited the formation and inflammation of subcutaneous fat, and two significantly reduced muscle mass gain and reduced dyslipidemia and liver steatosis [46]. Research indicates that a similar effect is not achieved by administering sodium butyrate, which suggests the action of microorganisms as factors inhibiting the development of obesity [46]. Research was undertaken to explain the effect of the CCFM1299 (C20_1_1) strain in preventing the development of obesity. For this purpose, CCFM1299 was orally administered to mice treated with a high-fat diet for 12 weeks. The results indicated that this strain inhibits the development of obesity by increasing energy expenditure. In addition, it increases the expression of genes related to the thermogenesis of brown adipose tissue (BAT). The term strain may also influence the expression of immune-related genes in epididymal white adipose tissue (eWAT). The immunomodulatory effect may be achieved by affecting the complement system, since factor D (CFD) gene expression was significantly decreased. The research results also indicate that the C. butyricum strain affects the metabolism of bile acids because increased concentrations of ursodeoxycholic acid (UDCA) in feces and taurohyodeoxycholic acid (THDCA) have been recorded in serum [47]. The presented results indicate the potential effective use of C. butyricum supplements in inhibiting the development of obesity [47].
One of the main complications of obesity is the development of diabetes. It is a chronic metabolic disease that can develop for a very long time and initially causes no apparent symptoms. It is associated with an increased glucose level in the blood due to the lack of insulin secretion, its reduced amount or tissue resistance to the action of insulin. If undiagnosed, untreated or treated incorrectly, it can cause damage to organs (eyes, heart, kidneys) and nerves.
Tayyib et al. examined the effects of Clostridium butyricum and magnesium supplementation on intestinal dysbiosis and blood sugar levels. The research was conducted on diabetic rats on an elemental diet. The control group received metformin, test group G1 received Clostridium butyricum (1.5 × 105 CFU/day), test group G2 received magnesium (500 mg/kg/day), and the study group (G3) received Clostridium butyricum (1.5 × 105 CFU/day) and magnesium (300 mg/kg/day). Blood glucose and magnesium levels and a complete blood count were tested. Additionally, blood glucose levels were randomly monitored twice a week for three weeks. The results indicate that Clostridium butyricum effectively balanced blood glucose levels compared to those of other groups. Moreover, it restores the dysbiosis of microorganisms [48]. Therefore, it is justified to introduce C. butyricum into the diet of people with type 2 diabetes [48].
It is known that intestinal dysbiosis influences the development of diabetes and its complications, but the mechanism of these is only partially known. Zhou et al. studied the relationship between the gut microbiota and vascular inflammation in diabetic mice [49]. The research results indicate that the amount of CB in diabetic mice was significantly lower compared to that in the control group. Additionally, impaired vascular function, inflammation in arterial tissue, and increased retained oxygen species were demonstrated. It is worth noting that CB administration rebalanced the intestinal microflora and protected vascular function in diabetic mice by activating the Nrf2/HO-1 pathway. Therefore, administering CB to people with diabetes may alleviate vascular changes and improve the intestinal microbiome.
Research indicates the wide use of CBM 588 in improving our health after various surgical procedures. There are many treatment methods in which the intestinal microflora plays a significant role. For example, in patients undergoing hematopoietic stem cell transplantation (HSCT), the intestinal microbiota plays a vital role in further prognosis, transplant effectiveness and complications, and the occurrence of graft-versus-host disease (GVHD). It has been shown that the administration Clostridium butyricum MIYAIRI 588 (CBM588) as a live biotherapeutic agent is associated with maintaining a normal intestinal microflora in the early period after HSCT. However, alpha species diversity decreased significantly in patients not treated with CBM588, while β diversity shows that CBM588 did not change intestinal microflora structure 7–21 days after HSCT. It is worth emphasizing that patients who developed GVHD showed structural changes in the microbiota compared to before the transplant, which was recorded 14 days before the development of GVHD. Enterococcus numbers significantly predominated in GVHD patients after HSCT, and the Bacteroides population did not change. However, in patients who received CMB588, Enterococcus and Bacteroides were reduced [50]. The results of the presented studies suggest that preoperative administration of CBM588 effectively maintains the balance of the intestinal microflora and improves the effectiveness of treatment [50].
Immune checkpoint inhibitors (ICIs) are the standard treatment for patients with advanced non-small cell lung cancer (NSCLC). This modern therapy benefits patients in increased progression-free survival (PFS) and overall survival (OS). Nevertheless, some patients experience primary or secondary resistance to ICI treatment, the reasons for which are not entirely clear [51,52,53]. These are related to the tumor’s molecular characteristics, the epigenetic profile, or the gut microbiome [54,55,56].
Clinical preliminary studies have shown a positive correlation between Clostridium butyricum MIYAIRI 588 (CBM588) supplementation and the effectiveness of ICIs in NSCLC treatment. Paz Del Socorro et al. tested in mouse models whether the strain could enhance the immunogenicity of tumor-draining lymph nodes to overcome ICI resistance [57]. They showed that CMB 588 improves the effectiveness of ICI (anti-programmed cell death protein 1, aPD-1) [57]. It is dependent on the acquisition of a regulatory phenotype of intestinal phagocytes that limits intestinal damage and accumulation of immunosuppressive Ror+ Treg (Retinoic Orphan Receptor Treg) in tumor-infiltrating lymph nodes after PD-1 blockade [57]. The authors indicate that live CBM588 can suppress a subset of Rorγt+ Tregs in the colonic mucosa. Therefore, the enhanced response to PD-1 blockade in patients supplemented with the live biotherapeutic CBM588 may be supported by the more significant accumulation of immunosuppressive Rorγt+ Treg to the colon, which may contribute to a more immunogenic reprogramming of tumor-draining lymph nodes that are strategically placed to infiltrate the tumor. In addition, the lowered frequency of Rorγt+ Treg at the tumor-infiltrating lymph nodes is linked to microbiota-modulated tryptophan catabolism upon CBM588 treatment. Further, the CBM588-induced immunogenic conversion of the tumor-infiltrating lymph nodes is related to the Indoleamine 2,3-Dioxygenase 1/Inerleukine 10 (IDO1/IL-10) axis upon PD-1 blockade (IDO1 catalyzes the initial step in the degradation of tryptophan) [57].
What is more, it was observed by the authors that there is dysbiosis induced by anti-PD-1, and the beneficial impact of CBM588 on the effectiveness of PD-1 blockade is linked to lowered alpha diversity of the gut microbiota [57].
Saitsu et al. [58] also describe a case supporting polypectomy treatment with C. butyricum. It is known that polypectomy during pregnancy increases the risk of premature birth and even miscarriage. The study authors administered probiotics orally, including Clostridium butyricum and 17-alpha-hydroxyprogesterone caproate, to a 30-year-old pregnant woman in whom an approximately 40 mm cervical polyp was detected. As a result, the development of the polyp, which disappeared in the 28th week of pregnancy, was reversed. The patient gave birth to a healthy baby at term. Therefore, it can be concluded that probiotics can effectively prolong pregnancy in the case of pathology.


Table 1.
The role of Clostridium butyricum in human health and disease.
Table 1.
The role of Clostridium butyricum in human health and disease.
| Clostridium botulinum as a Health Support | Source |
|---|---|
| With oral administration, the composition of the normal gastrointestinal microflora is regulated by increasing the beneficial microflora and reducing harmful strains of microorganisms; improves digestion and the functioning of the digestive system. | [39] |
| Creates an unfavourable environment for pathogenic organisms mainly by producing butyric acid and adhering to human epithelial cells, creating a protective mucosal barrier. | [39] |
| Prevents post-antibiotic diarrhea. | [39] |
| LAB could synergistically enhance the probiotic functions of C. butyricum. | [40] |
| Administration of C. butyricum effectively restores the intestinal microbial balance after colonoscopy and contributes to faster recovery. | [41] |
| Negative correlation between C. butyricum content and the predisposition to the development of obesity, and potential effective use as supplements in inhibiting the development of obesity. | [45,46] |
| Effectively balances blood glucose, alleviates vascular changes, and improves the intestinal microbiome, suggesting support of type 2 diabetes treatment. | [47,48] |
| Improves the effectiveness of HSCT and maintains the balance of the intestinal microflora. | [49] |
| In non-small cell lung cancer patients, supports the effectiveness of treatment that uses inhibitors of immunological control points. | [56] |
| Supports polypectomy treatment. | [57] |
3.3. Pathogenicity and Threats of Clostridium butyricum
While non-toxic strains of Clostridium butyricum are commonly used as probiotics and supplements to help treat many conditions, other strains can cause pathological conditions such as botulism in infants or necrotizing enterocolitis in premature infants. Some strains may also cause harmful effects on the intestinal mucosa. Moreover, the toxin gene has been identified based on genome sequencing [59].
It is also known that pets can also be a source of infection, such as a turtle, which was the source of infection in two infants in Ireland who were diagnosed with botulism [8].
Another threat that some strains of C. butyricum may cause is necrotizing enterocolitis (NEC). Clinical symptoms of varying degrees of severity include bleeding from the gastrointestinal tract, ulceration, and, consequently, necrosis of the mucous membrane, abdominal distention, gas in the portal venous gas, and pneumatosis intestinalis [59]. The mechanism of NEC formation is still unclear despite many studies on the pathogenesis of NEC. It is known that the bacterium most frequently involved in the pathogenesis of NEC belongs to the Clostridium, and the first association between C. butyricum and NEC was described in 1977. Unfortunately, the occurrence of NEC is associated with high mortality (20–30%); what is more, survivors experience long-term complications. The diagnosis and prevention of NEC is still difficult because the clinical symptoms and markers of NEC are not specific, which makes it difficult to diagnose the disease correctly. NEC is a multifactorial disease, and the susceptibility of premature infants to the disease is still unclear. The occurrence of NEC is caused by factors such as immaturity of the premature infant, enteral nutrition, and intestinal dysbiosis, which leads to an imbalance between pro- and anti-inflammatory factors [60]. Studies indicate an association between NEC and intestinal colonization by C. butyricum, C. neonatale, or C. perfringens in premature infants [60].
C. butyricum has various properties and may have different health effects depending on the strain. Many studies indicate the probiotic and health-promoting effects of C. butyricum strains. Unfortunately, as in the case of other bacteria, some strains can have very unfavorable effects, even leading to death, especially in newborns and small children (Figure 2). Nevertheless, Clostridium is a part of the intestinal microflora, and understanding the relationships between other microorganisms and their impact on health is still a challenge for scientists.
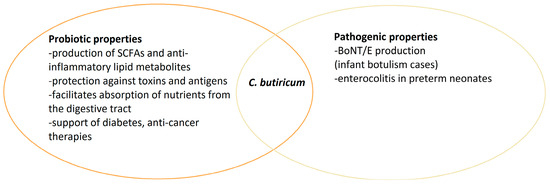
Figure 2.
Dual nature of C. butyricum species.
4. Genetic Mechanism of bont Genes Expression in Non-C. botulinum Strains
The botulinum toxin gene has a rather complex molecular structure. There is a fixed element is the NTNH (non-toxic, non-haemagglutinin) fragment. It plays a protective role for the toxin NTNH, protecting the toxin from the acid environment in the stomach [61,62].
Smith et al. [63] studied the structure, distribution, and gene sequence of eight different toxin complexes representing four different BoNT/A subtypes (BoNT/A1-Ba4) and one BoNT/B1 responsible for most cases of botulism in humans [63]. The strains representing these subtypes were Hall Sanger ATCC 3502, CLB A1 ATCC 19397, CLC A1 Hall, and pCLD B1 okra. The gene arrangement within the three BoNT/A1 strains and the BoNT/B1 okra strain is identical in the orientation and composition of the individual functional and structural elements. It includes NTNH, BoNT, HA70, HA17, HA33, and BotR, (A1 or B1) [63].
Strains representing the BoNT/A2, BoNT/A3, and BoNT/A4 subtypes (CLM A2 Kyoto-F, pCLK A3 Loch Maree, pCLJ A4, respectively) contain the orfX3, orfX2, orfX1, BotR, p47, ntht, and bont genes. In the pCLJ bvB strain, BoNT/tvB is located in the plasmid, approximately 97 kb away from BoNT/A4 [63]. The different types of bont clusters have characteristic flanking sequences, which are not irrelevant for the horizontal transfer of bont genes. Thus, in strain A1, we find IS3 and flagellin sequences. In strain A2 sequences, arsC, and A3 IS3, IS605, and lycA are also present in strain A4. IS256 is present in bvB and B1, where functional flagellin sequences are also present.
Clostridium botulinum is divided into four groups (I–IV), where groups I and II cause human diseases, and III cause disease in animals. Human cases associated with Group III are extremely rare. An analysis of the genomes of groups I, II, and III has shown that the toxin genes, including the bont cluster, are carried by plasmids. With Group III, they are found within prophages. The Group III genomes contain many plasmids carrying various toxin genes. Some genes are also found in Clostridium species other than C. botulinum; some move between different plasmids within the same type [64].
Clostridium botulinum group I strains mainly have botulinum neurotoxin genes on their chromosome, while some genes (bont/a, bont/b, and bont/f) are located on plasmids. Sometimes, these genes are found within the chromosome, and in other strains, they are located within plasmids. It extends to bont genes of the same subtype in some cases. These bont gene clusters’ varied locations illustrate the different phases of horizontal gene transfer and demonstrate that bont gene location is a fluid.
Nawrocki et al. [65] carried out studies on the transfer of botulinum toxin genes between strains of the Clostridium. A pCLJ, a 270 kb plasmid encoding two BoNTs, was transferred from C. botulinum 657Ba (an auxotrophic donor strain created by multiple chemical mutagenesis). The study demonstrated the transfer of a 270 kb pCLJ plasmid containing two bont genes from the donor strain to various Clostridium. The frequency of transfer was highest to other C. botulinum group I strains. The plasmid was also transferred to non-toxic Clostridium species, namely C. sporogenes and C. butyricum. Ng et al. [66] have analyzed the genomic organization and evolutionary relatedness in four closely related A1 or A1(B) types of C. botulinum strains. They carried out an analysis of both the core botulinum toxin cluster and the surrounding functional genes. The authors indicate 90% similarity of core genes and 96% similarity of functional gene groups in these four genomes. Matching the genomes of the three A1 strains revealed a very similar chromosome structure with three small gaps in the ATCC 19397 genome and one additional gap in the Hall A genome, suggesting that ATCC 19379 is an evolutionary intermediate relationship between Hall A and ATCC 3502. Four gap regions indicated potential horizontal gene transfer and recombination events necessary for the evolution of A1 strains [66]. The authors’ analysis of the nearest region downstream of the HA+ cluster (B) in NCTC 2916 suggested possible recombination between HA+ (HA+ cluster for producing neurotoxin complexes composed with hemagglutinins (HA), a non-hemagglutinin non-toxic (NTNH), and BoNT proteins) clusters located on the plasmid and chromosome [66]. BoNT/A1 strains are unique in that the bont/A1 genes may be found as part of either orfX+ or ha+ gene clusters with this toxin subtype. The bont/A1 genes that are part of the ha+ gene cluster were inserted into an existing bont/(B) gene cluster via homologous recombination within the bont/B ntnh gene, producing a hybrid B-A ntnh gene and inserting the bont/A gene within the bont/B gene cluster [67]. In the case of NCTC 2916 and other BoNT/A1(B) strains, the silent (B) gene follows the bont/A1 gene cluster, but in HA+ BoNT/A1 strains, the bont/B genes are absent. No plasmids are present in these strains, so if the original gene transfer occurred between a plasmid-borne bont/A1 gene cluster and the chromosomally located bont/B gene cluster, the plasmid was subsequently lost. However, this is more likely between chromosomally located orfX+ bont/A1 gene clusters and ha+ bont/B gene clusters coexisting within the same chromosome. The potential sequence equality between serotypes in the IS3, IS256, IS605, lycA, arsC, and flagellin regions is interesting, which may influence the opportunity and strength of horizontal gene transfer [68]. It can be based on qPCR genotyping of flaVR variable regions [69]. Woudsta et al. [69] point to genetic variability in flagellin that may be geographically specific, and they made these conclusions by studying strains isolated from European and Canadian cases. In their study, the authors point out the genetic diversity of flaVR among C. botulinum strains and the clustering of flaVR types into five significant subgroups. Subgroups 1, 3, and 4 harbor proteolytic Clostridium botulinum, subgroup 2 consists exclusively of non-proteolytic C. botulinum, and subgroup 5 is specific to E-type C. butyricum. These are the conclusions of a study published in 2013. The BoNT-producing bacteria group currently includes numerous non-clostridial species (Table 2). The variable region of flagellin was useful in a study by Valdezate et al. [70] published in 2023. They studied the genetic diversity and phylogenetic relationships of Clostridium botulinum from foodborne botulism and infant cases. The botulinum toxin gene subtype (bont), the variable region of the flagellin gene (flaVR), and the seven-gene multilocus sequence type were examined by sequencing 37 bacterial cultures. It is well-known that botulism due to BoNT/B2 is prevalent in several Western European countries. The surprising finding is that some of the BoNT/B2 strains in France and Italy are C. botulinum group I, and some are C. sporogenes [70].

Table 2.
Division based on physiological and genetic differences between C. botulinum and other Clostridium strains. The other strains of BoNT-like producing bacteria.
For each toxinotypes, the existence of bont genes in mobile genetic elements (except bont/C and bont/D) has been proven. Examples of subtypes BoNT/A2, BoNT/A3, BoNT/B1, BoNT/B2, BoNT/E1, BoNT/E3, and BoNT/E10 have also been described, in which the same botulinum clusters were demonstrated in the chromosome and in the plasmids. Extrachromosomal elements seem to be specific to certain metabolic groups of C. botulinum or other species of BoNT-producing Clostridia. For example, plasmids characteristic of C. botulinum group I (C. parabotulinum) are not significantly similar to plasmids of C. botulinum group II or C. argentinense, but also to sequences of C. botulinum group III (Clostridium novyi sensu lato) bacteriophages containing bont/C or bont/D. However, homology was observed between plasmids derived from C. parabotulinum and C. sporogenes. Smith et al. [71] conducted research using whole-genome sequencing (NGS), demonstrating that plasmids containing botulinum toxin genes can integrate into the bacterial chromosome, which may result in new strains of Clostridia stably producing BoNT [71]. The research was conducted using Clostridium sporogenes BoNT/B1 strain CDC 1632, C. argentinense BoNT/G strain CDC 2741, and Clostridium parabotulinum BoNT/B1 strain DFPST0006. Chromosomal bont gene clusters have been identified in the genomes of this bacteria in plasmid-like sequences or nested in large contigs, with no evidence of extrachromosomal elements [71]. The researchers demonstrate in the paper that full-length plasmid DNA carrying complete neurotoxin gene clusters has undergone integration into the chromosomes of three different bacterial species: C. parabotulinum, C. sporogenes, and C. argentinense. A fragment of the chromosomal sequence identified in C. sporogenes shared 99.5% identity with bont/B1-containing plasmid pNPD7 of C. sporogenes CDC 67071 [71].
Further, CDC 2741 contig AYSO01000020 contained a ~140 kb region, which shared 99.99% identity with plasmid pRSJ17_1 of C. argentinense BoNT/G strain 89G. At least DFPST0006 contig JACBDK0100002 contained a region that shared 100% identity with the bont/B1-containing plasmid pCLD of C. parabotulinum Okra. These studies show that not only the horizontal transfer of bont genes is vital for BoNT toxin production by strains other than C. botulinum but also the mechanisms that allow the integration of bont genes into their bacterial chromosome. It is an essential issue because integration into the chromosome can lead to a stable genetic construct, which strongly facilitates the identification by molecular biology methods of the presence of botulinum toxin-production genes. Plasmids carrying botulinum toxin genes can be temporarily lost; therefore, despite the occurrence of botulism’s clinical symptoms, identifying the cause of its onset is extremely difficult. On the other hand, the stable integration of bont-carrying plasmid fragments into the chromosome due to the biohazard of strains of the genus Clostridium, which are considered harmless and non-poisonous, seems to be a rather worrying phenomenon [71].
The other side of the BoNT story is that botulinum toxin is used in medicine, the pharmaceutical industry, and cosmetology. Despite being the most potent of the known biological toxins, it has applications in the treatment of spastic conditions, salivation, and neurological conditions, and is also used in oncological treatment [72]. Structural studies of BoNTs are critical. Safety in BoNT research is necessary for public health risk management, food preservation strategy development, and understanding toxinogenesis.
5. Non-Clostridium BoNT-like Producing Strains
The bont-like gene cluster has also been observed in Enterococcus commensal bacteria in the gastrointestinal tract, both Enterococcus faecium and Enterococcus faecalis [9,73,74,75].
It has been proven that the gene cluster encoding BoNT/En (eBoNT/J) could be located on the conjugation plasmid of Enterococcus faecium, and BoNT/En cleaves both VAMP2 and SNAP-25 required for synaptic transmission in neurons, but differs from the sites of other known BoNTs [73]. BoNT/En does not appear to be toxic to mice. However, a chimeric toxin composed of the H chain of BoNT/A and the L chain of BoNT/En leads to paralysis. It induces symptoms of botulism, suggesting that this putative BoNT may have a different mechanism of action. The authors point out that the ability of common commensal strains of the Enterococcus genus to acquire genes that allow the production of botulinum toxin is highly dangerous, and the possibility of their emergence in multidrug-resistant strains appears to threaten biosafety, especially since E. faecium is responsible for multidrug-resistant hospital infections [73].
Additionally, Brunt et al. [9] identified the bont-like gene cluster in Enterococcus. They identified (by bioinformatics tools) and described a novel bont gene cluster from Enterococcus sp. 3G1_DIV0629, with a typical ntnh gene and an uncommon orfX arrangement. The sequence of this gene cluster shows that its closest relative is the bont/X cluster from the C. botulinum 111 strain. The amino acid sequence homology with BoNT/X is only nearly 39%. Still, modelling the 3D structure shows that the putative eBoNT/J is very similar to the neurotoxin BoNT/A structure. The authors indicate that further work is needed to investigate whether this structural variation will have important implications for the potential use of the putative eBoNT/J as a therapeutic agent [9].
Tehran et al. [74], in their review, point out that botulinum toxins are produced by bacteria other than Clostridium botulinum, which offers new opportunities for research in both the pharmaceutical and medical fields. Botulinum toxins produced by bacteria of the Enterococcus genus, as well as other non-clostridium bacteria that produce BoNT, may differ in structure and action [74].
In addition to BoNT-producing clostridia homologs, other taxa distinct from this genus have been identified as carrying the botulinum-like gene cluster. Such as Chryseobacterium piperi (bont/Cp1), Enterococcus faecium, or Weissella oryzae. However, the production of botulinum toxin by strains other than Clostridium has not been demonstrated. The discoveries mentioned above further complicate the taxonomic division of BoNT-producing bacteria [76].
Poulain et al. [75] note that the topic of the production of botulinum toxins—a very diverse group of toxins by bacteria, including those other than Clostridium botulinum—is still open. The latest molecular biology technologies have dramatically accelerated the work on understanding the action of BoNT and the spread of its genes between microorganisms. However, the subject is still topical, inexhaustible, and requires much research [75].
6. Problem with Classifications and Taxonomy
Different metabolic and physiological features cause the designation of bacteria able to produce botulinum toxins or carrying botulinum genes cluster highly problematic. Including all microbiological strains carrying genes that determine the production of botulinum toxins into a common framework is impossible. This phenotypic and genetic diversity is why classifying microorganisms containing the botulinum genes cluster has undergone many changes. Additionally, the mentioned bont gene cluster could be present in strains not taxonomically defined as Clostridium spp. This problem with taxonomic division does not exist in the case of C. tetani strains (it is known that both toxins, botulinum and tetanus, have the same typical ancestral toxin production-determining gene). In this case, the ability to produce the tetanus neurotoxin is specific and limited only to the mentioned species. Historically, BoNT-producing clostridia are defined as C. botulinum species, which, based on many discoveries made using biochemical and genetic tools, is not a correct definition and cannot be applied to all mentioned strains of this type. One of the earliest observations and division of the BoNT–producing clostridia was based on metabolic properties. It was noticed that some strains are proteolytic and non-proteolytic and that these strains differ from each other in lecithinase and lipase activity observed on egg yolk agar. On this basis, a division of the four metabolic groups was established. This grouping system was implemented to recognize the groups’ metabolic properties but did not make legitimate taxonomic changes. Moreover, this system showed that four distinct taxons should be highlighted. Over time, strains from species considered saprophytic (C. baratii, C. sporogenes) and even probiotic (C. butyricum) that were capable of producing botulinum toxin were discovered [1,2,76,77].
Genetic-based classification methods have evolved from DNA:DNA hybridization for 16S rRNA sequence analysis through pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), amplified fragment length polymorphism (AFLP), up to whole-genome sequence analysis enabling average nucleotide identity (ANI) and single-nucleotide polymorphism (SNP) comparisons. These techniques proved that the above-mentioned groups could be confirmed as distinct species based on metabolic features [76].
The taxonomic changes of Clostridium spp. are still in progress (http://www.bacterio.net, accessed on 12 July 2024) [78,79].
The reclassification of Clostridium strains capable of producing botulinum toxin is still under discussion [79,80].
A new recommendation for taxonomic division was proposed by Smith et al. [79]. They proposed Latin binomial names for all members of each metabolic group. According to the authors, the new taxonomic division should include the following species: Proteolytic group I of C. botulinum should be changed to Clostridium parabotulinum; the designation of non-proteolytic group II should be changed to C. botulinum; the proposal for group III assumes changing the name to “C. novyi sensu lato”, closely related to C. novyi [60]. BoNT-producing clostridia’s remaining names remain (C. argentinense, C. baratii, C. butyricum, and C. sporogenes) [3]. This reclassification is not associated with the production of botulinum toxins. BoNT-producing strains can be distinguished by type or subtype classification, e.g., C. parabotulinum BoNT A1 or “C. baratii BoNT F” [79,81].
Bont-like genes are found not only in members of Clostridium spp. but also in other non-Clostridium strains, which makes the taxonomic division even more confusing, and it is impossible to limit BoNT-producing Clostridia to the species C. botulinum. As suggested by Smith et al. [79], the division of Clostridium strains capable of producing botulinum toxin takes into account genetic, metabolic, and phenotypic diversity and the diversity of subtypes within individual groups, which enable differentiation according to the type of toxin produced. However, new genetic discoveries suggesting the possibility of botulinum-like cluster genes in bacteria other than Clostridia means that the division, according to Smith et al. [79], does not exhaust the classification possibilities. Thus, taxonomic divisions evolve continuously with discoveries. A major challenge seems to be to include microorganisms possessing bont or bont-like genes into the appropriate taxa, taking into account their phenotypic and genetic features. Moreover, the greatest challenge seems to be understanding the mechanisms conditioning the appearance of the mentioned genes in microorganisms considered saprophytic, which is associated with ensuring microbiological safety in the food chain.
7. Conclusions
Despite its historical definition, C. botulinum is not the only species capable of producing botulinum toxins. As time passes and molecular biology tools develop, reports indicate the possibility of producing toxins or the presence of a botulinum cluster in bacteria that were not classified as C. botulinum. These findings highlight the need for a new taxonomic classification that is adequate to the current state of knowledge. Despite the updates, the taxonomic division will continue to evolve, which also creates the need to thoroughly understand the mechanisms determining the interspecies transfer of the botulinum cluster, as well as the need to develop methods for detecting bacteria predisposed to the production of botulinum toxins.
Author Contributions
Conceptualization, T.G. and A.G.; writing—original draft preparation, A.G., T.G., A.J. and K.R.-T.; writing—review and editing, T.G. and A.G.; supervision, T.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Samul, D.; Worsztynowicz, P.; Leja, K.; Grajek, W. Beneficial and harmful roles of bacteria from the Clostridium genus. Acta Biochim. Pol. 2013, 60, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Erbguth, F.J. From poison to remedy: The chequered history of botulinum toxin. J. Neural Transm. 2008, 115, 559–565. [Google Scholar] [CrossRef]
- Ciccarelli, A.S.; Giménez, D.F. Cryoprotein produced by Clostridium botulinum type G. Infect. Immun. 1972, 5, 985–986. [Google Scholar] [CrossRef] [PubMed]
- Cato, E.P.; Stackebrandt, E. Taxonomy and Phylogeny. In Clostridia; Minton, N.P., Clarke, D.J., Eds.; Springer: Boston, MA, USA, 1989; pp. 1–26. ISBN 978-1-4757-9718-3. [Google Scholar]
- Carter, A.T.; Peck, M.W. Genomes, neurotoxins and biology of Clostridium botulinum Group I and Group II. Res. Microbiol. 2015, 166, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; East, A.K. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol. 1998, 84, 5–17. [Google Scholar] [CrossRef]
- Scalfaro, C.; Iacobino, A.; Grande, L.; Morabito, S.; Franciosa, G. Effects of megaplasmid loss on growth of neurotoxigenic Clostridium butyricum strains and botulinum neurotoxin type E expression. Front. Microbiol. 2016, 7, 217. [Google Scholar] [CrossRef]
- Brunt, J.; Carter, A.T.; Stringer, S.C.; Peck, M.W. Identification of a novel botulinum neurotoxin gene cluster in Enterococcus. FEBS Lett. 2018, 592, 310–317. [Google Scholar] [CrossRef]
- Truong, R.D.; Do, V.A.; Njaravelil, K.A.; Ayesu, K.; Madruga, M.; Carlan, S.J. Unusual Case of Ludwig Angina Caused by Clostridium sporogenes in an Immunocompromised HIV-Positive Patient with Alcoholism and Dental Abscess. Am. J. Case Rep. 2023, 24, e941731. [Google Scholar] [CrossRef]
- Lorenzo, J.M.; Munekata, P.E.; Dominguez, R.; Pateiro, M.; Saraiva, J.A.; Franco, D. Main Groups of Microorganisms of Relevance for Food Safety and Stability: General Aspects and Overall Description; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780128110324. [Google Scholar]
- Alataby, H.A.; Krishnamoorthy, V.; Ndzelen, L.; Kenne, F.M.; Valenti, K.; Savermuttu, J.; Nfonoyim, J. Clostridium Sporogenes Causing Bacteremia Originated from the Skin and Soft Tissue Infection in an Immunocompetent Patient—Case Report and Literature Review. Int. J. Crit. Care Emerg. Med. 2020, 6, 95. [Google Scholar] [CrossRef][Green Version]
- Vecchio, M.J.; Jankowich, M.; Qadir, H.; Gaitanis, M.; Menon, A. Cognitive Biases in the Era of COVID-19: A Case of Clostridium sporogenes Bacteremia in a Patient with Small Bowel Obstruction. Case Rep. Infect. Dis. 2020, 2020, 8812635. [Google Scholar] [CrossRef] [PubMed]
- Bodey, G.P.; Rodriguez, S.; Fainstein, V.; Elting, L.S. Clostridial bacteremia in cancer patients. A 12-year experience. Cancer 1991, 67, 1928–1942. [Google Scholar] [CrossRef] [PubMed]
- Abusnina, W.; Shehata, M.; Karem, E.; Koc, Z.; Khalil, E. Clostridium sporogenes bacteremia in an immunocompetent patient. IDCases 2019, 15, e00481. [Google Scholar] [CrossRef] [PubMed]
- Rechner, P.M.; Agger, W.A.; Mruz, K.; Cogbill, T.H. Clinical features of clostridial bacteremia: A review from a rural area. Clin. Infect. Dis. 2001, 33, 349–353. [Google Scholar] [CrossRef]
- Shen, D.X.; Babady, N.E.; Chen, R.; Gilhuley, K.; Tang, Y.W. Septicaemia caused by Clostridium sporogenes: Two case reports and a literature review. Rev. Res. Med. Microbiol. 2013, 24, 81–83. [Google Scholar] [CrossRef]
- Cobo, F.; Pérez-Carrasco, V.; García-Salcedo, J.A.; Navarro-Marí, J.M. Bacteremia caused by Clostridium sporogenes in an oncological patient. Rev. Esp. Quimioter. 2023, 36, 217–219. [Google Scholar] [CrossRef]
- Stabler, S.; Titécat, M.; Duployez, C.; Kipnis, E.; Dessein, R.; Le Guern, R. Clinical relevance of Clostridium bacteremia: An 8-year retrospective study. Anaerobe 2020, 63, 102202. [Google Scholar] [CrossRef]
- Corbett, C.E.; Wall, B.M.; Cohen, M. Empyema With Hydropneumothorax and Bacteremia Caused by Clostridium sporogenes. Am. J. Med. Sci. 1996, 312, 242–245. [Google Scholar] [CrossRef]
- Malmborg, A.S.; Rylander, M.; Selander, H. Case report: Primary thoracic empyema caused by clostridium sporogenes. Scand. J. Infect. Dis. 1970, 2, 155–156. [Google Scholar] [CrossRef]
- Inkster, T.; Cordina, C.; Siegmeth, A. Septic arthritis following anterior cruciate ligament reconstruction secondary to Clostridium sporogenes; a rare clinical pathogen. J. Clin. Pathol. 2011, 64, 820–821. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, C.R.; Demello, F.J.; Haglin, J.J. Gangrene infection: New approaches to an old disease. Surg. Clin. N. Am. 1975, 55, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Kanaujia, R.; Dahiya, D.; Banda, A.R.; Ray, P.; Angrup, A. Non-traumatic gas gangrene due to Clostridium sporogenes. Lancet Infect. Dis. 2020, 20, 754. [Google Scholar] [CrossRef] [PubMed]
- Sárvári, K.P.; Schoblocher, D. The antibiotic susceptibility pattern of gas gangrene-forming Clostridium spp. clinical isolates from South-Eastern Hungary. Infect. Dis. 2020, 52, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Hara-Kudo, Y.; Yamakawa, Y.; Kumagai, S. Purification and some properties of Clostridium sporogenes hemorrhagic toxin. Biochem. Biophys. Res. Commun. 1996, 227, 413–418. [Google Scholar] [CrossRef]
- Brunt, J.; van Vliet, A.H.M.; Carter, A.T.; Stringer, S.C.; Amar, C.; Grant, K.A.; Godbole, G.; Peck, M.W. Diversity of the Genomes and Neurotoxins of Strains of Clostridium botulinum Group I and Clostridium sporogenes Associated with Foodborne, Infant and Wound Botulism. Toxins 2020, 12, 586. [Google Scholar] [CrossRef]
- Williamson, C.H.D.; Sahl, J.W.; Smith, T.J.; Xie, G.; Foley, B.T.; Smith, L.A.; Fernández, R.A.; Lindström, M.; Korkeala, H.; Keim, P.; et al. Comparative genomic analyses reveal broad diversity in botulinum-toxinproducing Clostridia. BMC Genom. 2016, 17, 180. [Google Scholar] [CrossRef]
- Mazuet, C.; Legeay, C.; Sautereau, J.; Ma, L.; Bouchier, C.; Bouvet, P.; Popoff, M.R. Diversity of group i and II clostridium botulinum strains from France including recently identified subtypes. Genome Biol. Evol. 2016, 8, 1643–1660. [Google Scholar] [CrossRef]
- Smith, T.J.; Hill, K.K.; Raphael, B.H. Historical and current perspectives on Clostridium botulinum diversity. Res. Microbiol. 2015, 166, 290–302. [Google Scholar] [CrossRef]
- Giordani, F.; Fillo, S.; Anselmo, A.; Palozzi, A.M.; Fortunato, A.; Gentile, B.; Azarnia Tehran, D.; Ciammaruconi, A.; Spagnolo, F.; Pittiglio, V.; et al. Genomic characterization of Italian Clostridium botulinum group I strains. Infect. Genet. Evol. 2015, 36, 62–71. [Google Scholar] [CrossRef]
- McLauchlin, J.; Grant, K.A.; Little, C.L. Food-borne botulism in the United Kingdom. J. Public Health 2006, 28, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Franciosa, G.; Maugliani, A.; Scalfaro, C.; Aureli, P. Evidence that plasmid-borne botulinum neurotoxin type B genes are widespread among Clostridium botulinum serotype B strains. PLoS ONE 2009, 4, e4829. [Google Scholar] [CrossRef]
- Liberato, V.; Benevenuti, C.; Coelho, F.; Botelho, A.; Amaral, P.; Pereira, N.; Ferreira, T. Clostridium sp. as Bio-Catalyst for Fuels and Chemicals Production in a Biorefinery Context. Catalysts 2019, 9, 962. [Google Scholar] [CrossRef]
- Moțățăianu, A.; Șerban, G.; Andone, S. The Role of Short-Chain Fatty Acids in Microbiota-Gut-Brain Cross-Talk with a Focus on Amyotrophic Lateral Sclerosis: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 15094. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, X.; Wang, Y.; Wang, T.; Fang, D.; Hu, K. Effects of Clostridium butyricum on Intestinal Microflora and Metabolism of Eriocheir sinensis. Int. J. Mol. Sci. 2023, 24, 13784. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission Implementing Decision of 10 March 2014. Off. J. Eur. Union 2014, 588, 30–33. [Google Scholar]
- Ariyoshi, T.; Hagihara, M.; Takahashi, M.; Mikamo, H. Effect of Clostridium butyricum on Gastrointestinal Infections. Biomedicines 2022, 10, 483. [Google Scholar] [CrossRef]
- Seki, H.; Shiohara, M.; Matsumura, T.; Miyagawa, N.; Tanaka, M.; Komiyama, A.; Kurata, S. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr. Int. 2003, 45, 86–90. [Google Scholar] [CrossRef]
- Hagihara, M.; Kuroki, Y.; Ariyoshi, T.; Higashi, S.; Fukuda, K.; Yamashita, R.; Matsumoto, A.; Mori, T.; Mimura, K.; Yamaguchi, N.; et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience 2020, 23, 100772. [Google Scholar] [CrossRef]
- So, J.S.; Oh, K.; Shin, Y. Growth stimulation of Clostridium butyricum in the presence of Lactobacillus brevis JL16 and Lactobacillus parabuchneri MH44. Food Sci. Technol. Braz. 2022, 42, 1–8. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, L.; Liu, J.; Kong, J.; Deng, X.; Guo, X.; Shan, J.; Zhou, D.; Li, W.; Lin, Y.; et al. Gut microbiota dynamics and fecal SCFAs after colonoscopy: Accelerating microbiome stabilization by Clostridium butyricum. J. Transl. Med. 2024, 22, 222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Z.; Zhang, X.; Lu, C.; Yang, W.; Xie, X.; Xin, H.; Lu, X.; Ni, M.; Yang, X.; et al. Effects of dietary Clostridium butyricum and rumen protected fat on meat quality, oxidative stability, and chemical composition of finishing goats. J. Anim. Sci. Biotechnol. 2024, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yun, Y.; Lai, Z.; Ji, S.; Yu, G.; Xie, Z.; Zhang, H.; Zhong, X.; Wang, T.; Zhang, L. Supplemental Clostridium butyricum modulates lipid metabolism by reshaping the gut microbiota composition and bile acid profile in IUGR suckling piglets. J. Anim. Sci. Biotechnol. 2023, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Ma, J.; Lin, H.; Zhou, S.; Long, Z.; Qin, H.; Lin, Y.; Liu, L.; Huang, Z.; Li, Z. Analyzing the influence of Clostridium butyricum on liver health in spotted sea bass (Lateolabrax maculatus) via transcriptomics and metabolomics. Aquac. Int. 2024, 32, 4717–4736. [Google Scholar] [CrossRef]
- Liao, J.; Liu, Y.; Pei, Z.; Wang, H.; Zhu, J.; Zhao, J.; Lu, W.; Chen, W. Clostridium butyricum Reduces Obesity in a Butyrate-Independent Way. Microorganisms 2023, 11, 1292. [Google Scholar] [CrossRef]
- Liao, J.; Liu, Y.; Yao, Y.; Zhang, J.; Wang, H.; Zhao, J.; Chen, W.; Lu, W. Clostridium butyricum Strain CCFM1299 Reduces Obesity via Increasing Energy Expenditure and Modulating Host Bile Acid Metabolism. Nutrients 2023, 15, 4339. [Google Scholar] [CrossRef]
- Tayyib, H.M.U.; Ali, A.; Jabeen, S.; Habib-ur-Rehman; Kamran, H.; Bajaber, M.A.; Usman, M.; Zhang, X. Restoration of gut dysbiosis through Clostridium butyricum and magnesium possibly balance blood glucose levels: An experimental study. BMC Microbiol. 2024, 24, 105. [Google Scholar] [CrossRef]
- Zhou, T.; Qiu, S.; Zhang, L.; Li, Y.; Zhang, J.; Shen, D.; Zhao, P.; Yuan, L.; Zhao, L.; Duan, Y.; et al. Supplementation of Clostridium butyricum Alleviates Vascular Inflammation in Diabetic Mice. Diabetes Metab. J. 2024, 48, 390–404. [Google Scholar] [CrossRef]
- Fukushima, K.; Kudo, H.; Oka, K.; Hayashi, A.; Onizuka, M.; Kusakabe, S.; Hino, A.; Takahashi, M.; Takeda, K.; Mori, M.; et al. Clostridium butyricum MIYAIRI 588 contributes to the maintenance of intestinal microbiota diversity early after haematopoietic cell transplantation. Bone Marrow Transplant. 2024, 59, 795–802. [Google Scholar] [CrossRef]
- West, H.; McCleod, M.; Hussein, M.; Morabito, A.; Rittmeyer, A.; Conter, H.J.; Kopp, H.-G.; Daniel, D.; McCune, S.; Mekhail, T.; et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 924–937. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Sosman, J.A.; Atkins, M.B.; Leming, P.D.; et al. Five-Year Survival and Correlates among Patients with Advanced Melanoma, Renal Cell Carcinoma, or Non-Small Cell Lung Cancer Treated with Nivolumab. JAMA Oncol. 2019, 5, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Boyero, L.; Sánchez-Gastaldo, A.; Alonso, M.; Noguera-Uclés, J.F.; Molina-Pinelo, S.; Bernabé-Caro, R. Primary and Acquired Resistance to Immunotherapy in Lung Cancer: Unveiling the Mechanisms Underlying of Immune Checkpoint Blockade Therapy. Cancers 2020, 12, 3729. [Google Scholar] [CrossRef]
- Kovács, S.A.; Győrffy, B. Transcriptomic datasets of cancer patients treated with immune-checkpoint inhibitors: A systematic review. J. Transl. Med. 2022, 20, 249. [Google Scholar] [CrossRef] [PubMed]
- Vu, S.H.; Vetrivel, P.; Kim, J.; Lee, M.-S. Cancer Resistance to Immunotherapy: Molecular Mechanisms and Tackling Strategies. Int. J. Mol. Sci. 2022, 23, 10906. [Google Scholar] [CrossRef] [PubMed]
- Paz Del Socorro, T.; Oka, K.; Boulard, O.; Takahashi, M.; Poulin, L.F.; Hayashi, A.; Chamaillard, M. The biotherapeutic Clostridium butyricum MIYAIRI 588 strain potentiates enterotropism of Rorγt+Treg and PD-1 blockade efficacy. Gut Microbes 2024, 16, 2315631. [Google Scholar] [CrossRef]
- Saitsu, Y.; Yoneda, S.; Fukuta, K. Management of a pregnant woman with a large cervical polyp and moderate genital bleeding in the first trimester. BMJ Case Rep. 2024, 17, e258163. [Google Scholar] [CrossRef]
- Cassir, N.; Benamar, S.; La Scola, B. Clostridium butyricum: From beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 2016, 22, 37–45. [Google Scholar] [CrossRef]
- Ferraris, L.; Balvay, A.; Bellet, D.; Delannoy, J.; Maudet, C.; Larcher, T.; Rozé, J.C.; Philippe, C.; Meylheuc, T.; Butel, M.J.; et al. Neonatal necrotizing enterocolitis: Clostridium butyricum and Clostridium neonatale fermentation metabolism and enteropathogenicity. Gut Microbes 2023, 15, 2172666. [Google Scholar] [CrossRef]
- Oguma, K.; Inoue, K.; Fujinaga, Y.; Yokota, K.; Watanabe, T.; Ohyama, T.; Takeshi, K.; Inoue, K. Structure and Function of Clostridium Botulinum Progenitor Toxin. J. Toxicol. Toxin Rev. 1999, 18, 17–34. [Google Scholar] [CrossRef]
- Gu, S.; Jin, R. Assembly and Function of the Botulinum Neurotoxin Progenitor Complex. Curr. Top. Microbiol. Immunol. 2013, 364, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Hill, K.K.; Foley, B.T.; Detter, J.C.; Munk, A.C.; Bruce, D.C.; Doggett, N.A.; Smith, L.A.; Marks, J.D.; Xie, G.; et al. Analysis of the Neurotoxin Complex Genes in Clostridium botulinum A1-A4 and B1 Strains: BoNT/A3, /Ba4 and /B1 Clusters Are Located within Plasmids. PLoS ONE 2007, 2, e1271. [Google Scholar] [CrossRef] [PubMed]
- Skarin, H.; Segerman, B. Horizontal gene transfer of toxin genes in Clostridium botulinum. Mob. Genet. Elem. 2011, 1, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.M.; Bradshaw, M.; Johnson, E.A. Botulinum neurotoxin-encoding plasmids can be conjugatively transferred to diverse clostridial strains. Sci. Rep. 2018, 8, 3100. [Google Scholar] [CrossRef] [PubMed]
- Ng, V.; Lin, W.-J. Comparison of assembled Clostridium botulinum A1 genomes revealed their evolutionary relationship. Genomics 2014, 103, 94–106. [Google Scholar] [CrossRef][Green Version]
- Hill, K.K.; Xie, G.; Foley, B.T.; Smith, T.J.; Munk, A.C.; Bruce, D.; Smith, L.A.; Brettin, T.S.; Detter, J.C. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol. 2009, 7, 66. Available online: https://bmcbiol.biomedcentral.com/articles/10.1186/1741-7007-7-66 (accessed on 28 August 2024). [CrossRef]
- Paul, C.J.; Twine, S.M.; Tam, K.J.; Mullen, J.A.; Kelly, J.F.; Austin, J.W.; Logan, S.M. Flagellin Diversity in Clostridium botulinum Groups I and II: A New Strategy for Strain Identification. Appl. Environ. Microbiol. 2007, 73, 2963–2975. [Google Scholar] [CrossRef]
- Woudstra, C.; Lambert, D.; Anniballi, F.; De Medici, D.; Austin, J.; Fach, P. Genetic Diversity of the Flagellin Genes of Clostridium botulinum Groups I and II. Appl. Environ. Microbiol. 2013, 79, 3926–3932. [Google Scholar] [CrossRef]
- Valdezate, S.; Carrasco, G.; Medina, M.J.; Garrido, N.; Del Pino, S.; Valiente, M.; Pallarés, M.P.; Villalon, P. Exploring the genetic background of the botulism neurotoxin BoNT/B2 in Spain. Microbiol. Spectr. 2023, 11, e0238023. [Google Scholar] [CrossRef]
- Smith, T.J.; Tian, R.; Imanian, B.; Williamson, C.H.D.; Johnson, S.L.; Daligault, H.E.; Schill, K.M. Integration of Complete Plasmids Containing Bont Genes into Chromosomes of Clostridium parabotulinum, Clostridium sporogenes, and Clostridium argentinense. Toxins 2021, 13, 473. [Google Scholar] [CrossRef]
- Grenda, T.; Grenda, A.; Krawczyk, P.; Kwiatek, K. Botulinum toxin in cancer therapy—Current perspectives and limitations. Appl. Microbiol. Biotechnol. 2022, 106, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lebreton, F.; Mansfield, M.J.; Miyashita, S.-I.; Zhang, J.; Schwartzman, J.A.; Tao, L.; Masuyer, G.; Martínez-Carranza, M.; Stenmark, P.; et al. Identification of a Botulinum Neurotoxin-like Toxin in a Commensal Strain of Enterococcus faecium. Cell Host Microbe 2018, 23, 169–176.e6. [Google Scholar] [CrossRef]
- Tehran, D.A.; Pirazzini, M. Novel Botulinum Neurotoxins: Exploring Underneath the Iceberg Tip. Toxins 2018, 10, 190. [Google Scholar] [CrossRef]
- Poulain, B.; Popoff, M.R. Why Are Botulinum Neurotoxin-Producing Bacteria So Diverse and Botulinum Neurotoxins So Toxic? Toxins 2019, 11, 34. [Google Scholar] [CrossRef]
- Grenda, T.; Jarosz, A.; Sapała, M.; Stasiak, K.; Grenda, A.; Domaradzki, P.; Kwiatek, K. Molecular Diversity of BoNT-Producing Clostridia—A Still-Emerging and Challenging Problem. Diversity 2023, 15, 392. [Google Scholar] [CrossRef]
- Hayashi, H.; Sakamoto, M.; Kitahara, M.; Benno, Y. Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol. Lett. 2006, 257, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Rainey, F.A. Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 2016, 66, 1009–1016. [Google Scholar] [CrossRef]
- Smith, T.; Williamson, C.H.D.; Hill, K.; Sahl, J.; Keim, P. Botulinum Neurotoxin-Producing Bacteria. Isn’t It Time that We Called a Species a Species? mBio 2018, 9, e01469-18. [Google Scholar] [CrossRef]
- Peck, M.W. Biology and Genomic Analysis of Clostridium botulinum. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2009; Volume 55, pp. 183–320. [Google Scholar]
- Suen, J.C.; Hatheway, C.L.; Steigerwalt, A.G.; Brenner, D.J. Genetic confirmation of identities of neurotoxigenic Clostridium baratii and Clostridium butyricum implicated as agents of infant botulism. J. Clin. Microbiol. 1988, 26, 2191–2192. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).