Abstract
CRISPR-Cas systems are adaptive immune mechanisms present in most prokaryotes that play an important role in the adaptation of bacteria and archaea to new environments. Shewanella algae is a marine zoonotic pathogen with worldwide distribution, which accounts for the majority of clinical cases of Shewanella infections. However, the characterization of Shewanella algae CRISPR-Cas systems has not been well investigated yet. Through whole genome sequence analysis, we characterized the CRISPR-Cas systems in S. algae. Our results indicate that CRISPR-Cas systems are prevalent in S. algae, with the majority of strains containing the Type I-F system. This study provides new insights into the diversity and function of CRISPR-Cas systems in S. algae and highlights their potential role in the adaptation and survival of these marine pathogens.
1. Introduction
The CRISPR-Cas system (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein) is a mechanism for natural immunity in bacteria, helping them to combat invading viruses (phages) by serving as adaptive immune systems that confer heritable resistance to foreign nucleic acids in most prokaryotes [1] and some viruses [2]. The CRISPR-Cas system stores and utilizes short DNA fragments that have been previously invaded by bacteriophages as immune memories, enabling bacteria to recognize and neutralize future invaders. This adaptive immune system allows bacteria to swiftly adapt to novel threats in their environment [3,4].
CRISPR-Cas systems have the ability to integrate segments of invading DNA as spacers into the CRISPR loci. Designed as a genome-editing tool, CRISPR–Cas systems require only a short RNA sequence and can recognize different target sequences [5]. Currently, CRISPR–Cas systems comprise two classes (class 1 and class 2) and are further classified into at least six different types [6]. According to the current classification, Class 1 encodes multiple Cas subunits to form a complex system, and Class 2 systems consist of a single multidomain protein and have been widely used as genome-editing tools, such as Cas9 and Cas12a [7,8].
Shewanella is a marine zoonotic pathogen with worldwide distribution. The predominant clinical infections it causes include blood stream infections from hepatobiliary sources and skin and soft tissue infections following seawater exposure [9,10].
Several highly diverse Shewanella species include S. algae, S. haliotis, S. putrefaciens, and S. xiamenensis [11,12]. Shewanella algae (the most common) and S. haliotis account for most clinical cases of Shewanella infections in the Western Pacific region [9,13]. Environment surveillance has also demonstrated these microorganisms are widespread in the marine environment, including water and aquaculture [14,15,16].
To date, a few genome analyses of the genus Shewanella have attempted to elucidate the association of genetic characteristics with potential pathogenic lineages [17,18,19]. There are two studies that have shown the presence of a CRISPR-Cas system in a single strain of S. putrefaciens [20] and S. xiamenensis [21]. However, the characterization of Shewanella using CRISPR-Cas systems has not yet been investigated, particularly for S. algae.
In this article, we present a whole-genome sequence analysis of S. algae and S. haliotis. We used a combination of molecular biology techniques and bioinformatics to investigate the CRISPR-Cas systems in S. algae and S. haliotis and to identify candidate virulence genes. By using a multiple-database-based approach, we were able to cross-validate our results and achieve a comprehensive understanding of these adaptive immune systems in the marine zoonotic pathogens.
2. Materials and Methods
2.1. Shewanella Strains
A total of 17 Shewanella strains were included in the study (Table 1). These strains were collected from both clinical and environmental sources, and included ten S. algae and seven S. haliotis (labeld as S. algae/S. haliotis), a closely-related strain of S. algae. The seven strains were initially classified as Shewanella haliotis and later changed to Shewanella algae by NCBI due to the publication by Szeinbaum et al. [22]. All 11 clinical isolates are associated with invasive infection. The strains were preliminary identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (bioMérieux, Marcy-l′Étoile, France) and Sanger sequencing of the 16S rRNA gene [23]. The primers used for amplification of the 16S rRNA gene were B27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and U1492R (5′-GGTTACCTTGTTACGACTT-3′). The PCR product was sequenced against the bacterial 16S rRNA gene sequences in the GenBank database of the National Center for Biotechnology Information using the BLASTn (optimized for Megablast) algorithm [24].

Table 1.
Shewanella strains in this study.
2.2. Whole-Genome Sequencing, Assembly, and Annotation
The strains were stored at −80 °C in Müller–Hinton broth containing 8.7% (vol/vol) glycerol until further usage. DNA was extracted from one colony of each strain grown on Trypticase Soy Agar with 5% sheep blood (Becton–Dickinson, Franklin Lakes, NJ, USA) using the QIAGEN Genomic-tip 100/G kit and the Genomic DNA Buffer Set (QIAGEN, Hilden, Germany) [25]. The DNA concentration was measured using a Qubit dsDNA HS Assay kit using Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). The genomic DNA was fragmented using Covaris S2 (Covaris, Woburn, MA, USA). The fragmented DNA was used to construct indexed PCR-free libraries using the multiplexed high-throughput sequencing TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA, USA), following the protocols provided by the manufacturer [26,27]. Whole-genome shotgun sequencing was performed using 2 × 250 bp paired-end sequencing on a MiSeq platform (Illumina, San Diego, CA, USA).
The reads were filtered using duk (http://duk.sourceforge.net/ (accessed on 10 August 2023)) and trimmed with the FASTQX-toolkit fastqTrimmer (https://github.com/agordon/fastx_toolkit (accessed on 10 August 2023)). The reads were then assembled using Velvet v. 1.2.07 [28] and the resulting contigs were scaffolded using ALLPATHS v. R46652 [29]. The assembled DNA sequences of the studied strains were annotated using the National Center for Biotechnology Information (NCBI) Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP). Functional classification of these annotated genes (e-value < 0.001) was carried out with the RPSBLAST v. 2.2.15 [30] in conjunction with the database of Clusters of Orthologous Groups of proteins (COGs). The sequencing data for this study have been uploaded in Supplementary Table S1 for reference.
2.3. CRISPR-Cas Systems Analysis
A multiple-database-based approach was used to analyze the CRISPR-Cas systems in the tested Shewanella strains. All predicated coding regions of the genome were subjected to analysis against CrisprCasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index (accessed on 13 September 2023)) [31], CRISPRminer (http://www.microbiome-bigdata.com/CRISPRminer/ (accessed on 14 September 2023)) [32], and CRIS-PRcasIdentifier (https://github.com/BackofenLab/CRISPRcasIdentifier (accessed on 15 September 2023)) [33] to identify and cross-validate CRISPR-Cas types. These software and websites were used to provide consistent results, but each one also provided additional information. For example, CrisprCasFinder additionally provides CRISPR spacers and repeat consensus [34], and CRISPRminer provides Phage alignment results.
In addition to identifying CRISPR-Cas systems, we also sought to identify candidate virulence genes in the Shewanella genomes. To facilitate this, we aligned the assembled genomes against the full Virulence Factors Database (VFDB) protein sequences dataset using BLASTX. We used the following criteria for identification: >45% identity; >450 bp aligned length; >95% alignment coverage; and E-value < 1 × 10−45.
2.4. Nucleotide Sequence Accession Number
The sequencing data for each isolate were submitted to GenBank with BioProject record number PRJNA312015.
3. Results
CRISPR-Cas systems were detected in all 17 strains in this study (Table 2). Analysis of the cas gene structures revealed that eight S. algae and five S. algae/S. haliotis genomes possessed a typical I-F cas structure, including the universal csy1, csy2, csy3, and cas6f genes as well as the type I signature genes cas3, which is fused to cas2, a unique feature of I-F-type systems. Furthermore, we identified both CAS-I-E and CAS-I-F in two closely related strains of Shewanella. The alignment results demonstrated the consistency in the distribution of cas genes for each Cas type. Two S. algae and two S. algae/S. haliotis genomes showed a type I-E structure, excluding the cas4 gene (Figure 1). All of the CRISPR-positive strains possessed only one valid CRISPR-Cas structure. The spacer counts ranged from 10 to 86 for type I-F and from 28 to 110 for type I-E systems. The CRISPR repeat sequences for each type are similar (Table 3).

Table 2.
Spacers and cas genes found in CRISPR-system in the study.
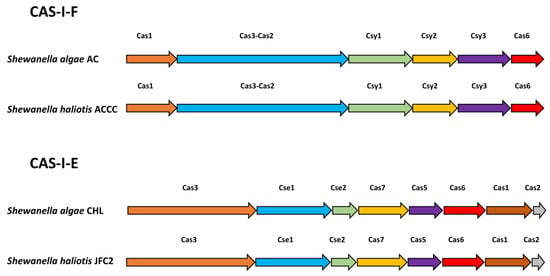
Figure 1.
Overview of Shewanella algae CRISPR-Cas structures.

Table 3.
Repeat consensus of Shewanella strains in the study.
Our analysis also revealed that 20 of the identified spacers had putative origins from phages. Most of these came from phages detected in Vibrio (Table 4). By aligning ORF-encoded protein sequences with the virulence factor database, we screened the Shewanella genome for putative virulence-associated genes. The genomic analysis revealed strain-specific gene tapW which is associated with twitching ATPase. S. algae also harbors exeG and gspG, which are associated with general secretion pathway protein G (Supplementary Table S2).

Table 4.
Phage-originated spacers in Shewanella algae.
4. Discussion
In this study, we aimed to conduct a sequence analysis and characterization of the CRISPR-Cas system in S. algae and S. haliotis. Type I-F and type I-E CRISPR-Cas systems were identified in all of the Shewanella strains analyzed in our study. The majority of clinical isolates contained a type I-F system. The type I-F and type I-E CRISPR-Cas systems were hypothesized to comprise a bacterial defense mechanism against phage and other genetic elements [4]. Previous studies have reported the presence of type I CRISPR-Cas systems in other species of Shewanella, including type I-F in S. putrefaciens [20] and type I-E in S. xiamenensis [21]. A further comparative genome analysis of 41 genomes supported these previous reports; the I-F type was reported to be the most frequent system (27/41; 65.85%) in S. algae, and S. putrefaciens and the type I-E system was found in S. xiamenensis strains (21.9%) [19].
Regarding the CRISPR arrays associated with each cas operon, our data show that the cas operons are well conserved in tested strains, displaying two distinct cas gene profiles. Cas operons are clusters of genes found in some bacteria that contain genes related to the CRISPR-Cas system. These gene clusters may help maintain the host’s defenses’ functionality and were found to be relevant in a recent study [19]. On the other hand, we also observed diverse spacers, suggesting a high diversity of phage environments and the potential immune function of the CRISPR-Cas system in Shewanella. S. algae is widely distributed in marine and freshwater habitats. An early study showed various spacers in different S. xiamenensis strains among CRISPR-Cas systems within changing environments [35].
Our study complements these findings and expands our understanding of CRISPR-Cas systems in S. algae. The development of the CRISPR field has led to many clinical and biological implications [36,37]. Genome editing based on the CRISPR-Cas system enables efficient and affordable gene engineering [38]. Active CRISPR systems allow for the transcription and processing of the CRISPR arrays into short CRISPR RNAs (crRNAs) that contain a spacer and are covalently linked to the Cas endonuclease [39]. When the cell is re-infected, this complex utilizes crRNA base complementarity to recognize and degrade DNA or RNA from elements containing the spacer sequence [40].
Previous research has shown that the spacers present in the CRISPR arrays of bacteria are acquired from phages that have invaded the bacteria [41]. This means that the CRISPR-Cas systems of bacteria maintain a record, or molecular memory, of previous infections. This is also the mechanism by which these systems confer adaptive immunity to bacteria and archaea against foreign DNA. Additionally, the presence of these spacers can also be used to trace the path of certain prokaryotic pathogens, as the spacers provide a record of the phages that have been present in a particular strain’s environment [42]. The fact that the spacers have originated from phages is crucial in understanding the biological records of past phage–bacteria interactions. Spacers can be utilized to predict hosts of unknown phages, providing insights into the history of interactions between phages and bacteria [43]. When studying the resistance of Shewanella spp., the number and types of spacer sequences in CRISPR also play an important role. One study showed that CRISPR/Cas9 genome editing technology can reverse the resistance of S. algae to carbapenem antibiotics [38]. The latest study indicated that spacer sequences are crucial components of the CRISPR system [35]. Different numbers and types of spacer sequences within various Shewanella-specific CRISPR systems demonstrate their significant role in the CRISPR-Cas system [35,44]. In general, S. algae were highly susceptible to aminoglycosides and carbapenems but were generally resistant to penicillin [45,46]. Reports of carbapenem and colistin resistance in S. algae have been increasing [38]. Therefore, it has been proposed as a potential source of antibiotic resistance in marine environments and deserves special attention in healthcare facilities.
Our findings of diverse spacers in S. algae and S. haliotis suggest a high diversity of phage environments and potential immune functions of CRISPR-Cas systems in these marine zoonotic pathogens. Other studies have also reported similar findings [35,47]. The bacterial resistance to phages and the activity of CRISPR was demonstrated to be correlated with CRISPR spacers diversity [48,49]. Previous spatially diverse models suggested that the CRISPR array evolved to have between 20 and 30 spacers [50], which is lower than our observation in Shewanella. Our results are also consistent with a coevolutionary model which suggested that the high diversity of spacers in these strains may have evolved in response to a diverse phage environment. These findings are in parallel with empirical observations of the adaptive immune system in bacteria [51]. The mechanisms of Shewanella immune system and the implications warrant further study [52].
Our study suggests that CRISPR-Cas systems are prevalent in S. algae and S. haliotis may play a role in their adaptation to new environments. The study further suggested a complex trade-off between gains and losses in CRISPR-Cas systems when adapting to new environments. There is growing evidence that CRISPR-Cas systems constrain horizontal gene transfer [53]. On the other hand, the elimination of immunity to newly acquired mobile elements that confer a fitness benefit could lead to the loss of CRISPR-Cas systems [54].
Further research is needed to determine the genetic diversity, pathogenicity, antibiotic resistance, and specific mechanisms of Shewanella’s immune system and the implications of these findings for understanding the ecology and pathogenesis of these organisms. Overall, our study expands our understanding of the diversity and function of CRISPR-Cas systems in Shewanella and creates new opportunities for investigating the role of these systems in the adaptation and survival of these pathogens in the marine environment.
5. Conclusions
In summary, our study found that CRISPR-Cas systems are prevalent in S. algae and S. haliotis, with most strains possessing a type I-F CRISPR-Cas system. Additionally, we identified a unique feature of a fused cas3 and cas2 gene in type I-F systems, and also identified different CRISPR-Cas systems in two different species of Shewanella. Furthermore, we found that most spacers had putative origins from phages and identified virulence-associated genes in the Shewanella genome. These findings provide new insights into the CRISPR-Cas systems in these marine zoonotic pathogens and the potential for these systems to play a role in their adaptation to new environments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13060439/s1, Table S1: Summary of sequencing and assembly data; Table S2: Virulence genes identified in the Shewanella.
Author Contributions
Conceptualization, Y.-T.H., C.-H.T. and C.-C.K.; methodology: J.-H.W., Y.-T.H. and C.-C.K.; software, Y.-T.H.; validation, J.-H.W., Y.-C.M. and C.-H.L.; formal analysis, J.-H.W. and T.-K.Y.; investigation, P.-T.H., Y.-C.M., C.-H.L., T.-K.Y. and C.-C.K.; data curation, P.-T.H., J.-H.W. and C.-H.T.; writing—original draft preparation, J.-H.W.; writing—review and editing, J.-H.W., C.-H.T., P.-T.H. and C.-C.K.; supervision, C.-H.T. and C.-C.K.; project administration, J.-H.W., C.-C.K. and C.-H.T.; funding acquisition, Y.-T.H., C.-H.T. and P.-T.H. All authors have read and agreed to the published version of the manuscript.
Funding
YTH was supported in part by the Ministry of Science and Technology (109-2221-E-194-038-MY3 and 111-2221-E-194-031-MY3); other specific grants from TCVGH (1133901E); and the Kaohsiung Armed Forces General Hospital (KAFGH_A_11001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequencing data for each isolate were submitted to GenBank with BioProject accession number PRJNA312015.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, S.; Liu, Y.; Koonin, E.V.; Severinov, K.; Prangishvili, D.; Krupovic, M. Virus-borne mini-CRISPR arrays are involved in interviral conflicts. Nat. Commun. 2019, 10, 5204. [Google Scholar] [CrossRef] [PubMed]
- Nunez, J.K.; Lee, A.S.; Engelman, A.; Doudna, J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 2015, 519, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Sashital, D.G. Mechanisms of Type I-E and I-F CRISPR-Cas Systems in Enterobacteriaceae. EcoSal Plus 2019, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb. Cell Fact. 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, J.; Wang, B.; Han, J.; Hao, Y.; Wang, S.; Ma, X.; Yang, S.; Ma, L.; Yi, L.; et al. Endogenous Type I CRISPR-Cas: From Foreign DNA Defense to Prokaryotic Engineering. Front. Bioeng. Biotechnol. 2020, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Lin, C.F.; Tung, K.C.; Shyu, C.L.; Wu, M.J.; Liu, J.W.; Chang, C.S.; Chan, K.W.; Huang, J.A.; Shi, Z.Y. Clinical and microbiological features of shewanella bacteremia in patients with hepatobiliary disease. Intern. Med. 2013, 52, 431–438. [Google Scholar] [CrossRef]
- Chen, Y.S.; Liu, Y.C.; Yen, M.Y.; Wang, J.H.; Wang, J.H.; Wann, S.R.; Cheng, D.L. Skin and soft-tissue manifestations of Shewanella putrefaciens infection. Clin. Infect. Dis. 1997, 25, 225–229. [Google Scholar] [CrossRef]
- Yu, K.; Huang, Z.; Li, Y.; Fu, Q.; Lin, L.; Wu, S.; Dai, H.; Cai, H.; Xiao, Y.; Lan, R.; et al. Establishment and Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for Detection of Shewanella Genus. Front. Microbiol. 2021, 12, 625821. [Google Scholar] [CrossRef] [PubMed]
- Hongyan, C.; Yujie, F.; Keyi, Y.; Zhenzhou, H.; Hang, D.; Duochun, W. Identification of Shewanella at species level based on16S rRNA and gyrB genes. Dis. Surveill. 2021, 36, 42–47. [Google Scholar] [CrossRef]
- Poovorawan, K.; Chatsuwan, T.; Lakananurak, N.; Chansaenroj, J.; Komolmit, P.; Poovorawan, Y. Shewanella haliotis associated with severe soft tissue infection, Thailand, 2012. Emerg. Infect. Dis. 2013, 19, 1019–1021. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Y.; Liu, P.Y.; Tseng, S.Y.; Lee, Y.H.; Ho, S.P. Characteristics and Phylogeny of Shewanella haliotis Isolated from Cultivated Shellfish in Taiwan. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 9895148. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.Y.; Liu, P.Y.; Lee, Y.H.; Wu, Z.Y.; Huang, C.C.; Cheng, C.C.; Tung, K.C. The Pathogenicity of Shewanella algae and Ability to Tolerate a Wide Range of Temperatures and Salinities. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 6976897. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Tung, K.-C.; Cheng, J.-F.; Wu, Z.-Y.; Chen, S.-Y.; Hong, Y.-K.; Huang, Y.-T.; Liu, P.-Y. Genomic characterization of carbapenem-resistant Shewanella algae isolated from Asian hard clam (Meretrix lusoria). Aquaculture 2019, 500, 300–304. [Google Scholar] [CrossRef]
- Lizarraga, W.C.; Mormontoy, C.G.; Calla, H.; Castaneda, M.; Taira, M.; Garcia, R.; Marin, C.; Abanto, M.; Ramirez, P. Complete genome sequence of Shewanella algae strain 2NE11, a decolorizing bacterium isolated from industrial effluent in Peru. Biotechnol. Rep. 2022, 33, e00704. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yu, K.; Fu, S.; Xiao, Y.; Wei, Q.; Wang, D. Genomic analysis reveals high intra-species diversity of Shewanella algae. Microb. Genom. 2022, 8, 000786. [Google Scholar] [CrossRef] [PubMed]
- Cerbino, G.N.; Traglia, G.M.; Ayala Nunez, T.; Parmeciano Di Noto, G.; Ramirez, M.S.; Centron, D.; Iriarte, A.; Quiroga, C. Comparative genome analysis of the genus Shewanella unravels the association of key genetic traits with known and potential pathogenic lineages. Front. Microbiol. 2023, 14, 1124225. [Google Scholar] [CrossRef]
- Dwarakanath, S.; Brenzinger, S.; Gleditzsch, D.; Plagens, A.; Klingl, A.; Thormann, K.; Randau, L. Interference activity of a minimal Type I CRISPR-Cas system from Shewanella putrefaciens. Nucleic Acids Res. 2015, 43, 8913–8923. [Google Scholar] [CrossRef]
- Wang, J.H.; Tseng, S.Y.; Tung, K.C. Genomic investigation of emerging zoonotic pathogen Shewanella xiamenensis. Ci Ji Yi Xue Za Zhi 2020, 32, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Szeinbaum, N.; Kellum, C.E.; Glass, J.B.; Janda, J.M.; DiChristina, T.J. Whole-genome sequencing reveals that Shewanella haliotis Kim et al. 2007 can be considered a later heterotypic synonym of Shewanella algae Simidu et al. 1990. Int. J. Syst. Evol. Microbiol. 2018, 68, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Syn, C.K.; Swarup, S. A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal. Biochem. 2000, 278, 86–90. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.L.; Jaszczyszyn, Y.; Thermes, C. Library preparation methods for next-generation sequencing: Tone down the bias. Exp. Cell Res. 2014, 322, 12–20. [Google Scholar] [CrossRef]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012, 22, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.; MacCallum, I.; Kleber, M.; Shlyakhter, I.A.; Belmonte, M.K.; Lander, E.S.; Nusbaum, C.; Jaffe, D.B. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 2008, 18, 810–820. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhao, S.; Ren, C.; Zhu, Y.; Zhou, H.; Lai, Y.; Zhou, F.; Jia, Y.; Zheng, K.; Huang, Z. CRISPRminer is a knowledge base for exploring CRISPR-Cas systems in microbe and phage interactions. Commun. Biol. 2018, 1, 180. [Google Scholar] [CrossRef] [PubMed]
- Padilha, V.A.; Alkhnbashi, O.S.; Shah, S.A.; de Carvalho, A.; Backofen, R. CRISPRcasIdentifier: Machine learning for accurate identification and classification of CRISPR-Cas systems. Gigascience 2020, 9, giaa062. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, J.; Yan, F.; Wang, G.; Li, Y.; Huang, J. CrisprVi: A software for visualizing and analyzing CRISPR sequences of prokaryotes. BMC Bioinform. 2022, 23, 172. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xia, F.; Xia, Y.; Li, J.; Hu, Y.; Deng, Y.; Zou, M. Pangenome analysis of Shewanella xiamenensis revealed important genetic traits concerning genetic diversity, pathogenicity and antibiotic resistance. BMC Genom. 2024, 25, 216. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Wang, C.Y.; Ko, W.C.; Hsueh, P.R. In vitro diagnostics of coronavirus disease 2019: Technologies and application. J Microbiol. Immunol. Infect. 2021, 54, 164–174. [Google Scholar] [CrossRef]
- Huang, W.H.; Teng, L.C.; Yeh, T.K.; Chen, Y.J.; Lo, W.J.; Wu, M.J.; Chin, C.S.; Tsan, Y.T.; Lin, T.C.; Chai, J.W.; et al. 2019 novel coronavirus disease (COVID-19) in Taiwan: Reports of two cases from Wuhan, China. J. Microbiol. Immunol. Infect. 2020, 53, 481–484. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Huang, Y.T.; Chao, W.C.; Ho, S.P.; Cheng, J.F.; Liu, P.Y. Reversal of carbapenem-resistance in Shewanella algae by CRISPR/Cas9 genome editing. J. Adv. Res. 2019, 18, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Moïra, B.D.; Pier-Luc, P.; Edwige, Z.; Shiraz, A.S.; Jacques, C.; Sylvain, M. Streamlining CRISPR spacer-based bacterial host predictions to decipher the viral dark matter. Nucleic Acids Res. 2021, 49, 3127–3138. [Google Scholar] [CrossRef] [PubMed]
- Corts, A.D.; Thomason, L.C.; Gill, R.T.; Gralnick, J.A. Efficient and Precise Genome Editing in Shewanella with Recombineering and CRISPR/Cas9-Mediated Counter-Selection. ACS Synth. Biol. 2019, 8, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Holt, H.M.; Gahrn-Hansen, B.; Bruun, B. Shewanella algae and Shewanella putrefaciens: Clinical and microbiological characteristics. Clin. Microbiol. Infect. 2005, 11, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Chia-Wei, L.; Cheng, J.F.; Tung, K.C.; Hong, Y.K.; Lin, J.H.; Lin, Y.H.; Tsai, C.A.; Lin, S.P.; Chen, Y.C.; Shi, Z.Y.; et al. Evolution of trimethoprim/sulfamethoxazole resistance in Shewanella algae from the perspective of comparative genomics and global phylogenic analysis. J. Microbiol. Immunol. Infect. 2022, 55, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Deem, M.W. Heterogeneous diversity of spacers within CRISPR (clustered regularly interspaced short palindromic repeats). Phys. Rev. Lett. 2010, 105, 128102. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, A.D.; Sun, C.L.; Plucinski, M.M.; Denef, V.J.; Thomas, B.C.; Horvath, P.; Barrangou, R.; Gilmore, M.S.; Getz, W.M.; Banfield, J.F. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput. Biol. 2012, 8, e1002475. [Google Scholar] [CrossRef]
- Tyson, G.W.; Banfield, J.F. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Env. Environ. Microbiol. 2008, 10, 200–207. [Google Scholar] [CrossRef]
- Haerter, J.O.; Sneppen, K. Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus. mBio 2012, 3, e00126-12. [Google Scholar] [CrossRef]
- Horvath, P.; Romero, D.A.; Coute-Monvoisin, A.C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef]
- Deem, M.W. CRISPR recognizes as many phage types as possible without overwhelming the Cas machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 7550–7552. [Google Scholar] [CrossRef]
- Wheatley, R.M.; MacLean, R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021, 15, 1420–1433. [Google Scholar] [CrossRef]
- Rollie, C.; Chevallereau, A.; Watson, B.N.J.; Chyou, T.Y.; Fradet, O.; McLeod, I.; Fineran, P.C.; Brown, C.M.; Gandon, S.; Westra, E.R. Targeting of temperate phages drives loss of type I CRISPR-Cas systems. Nature 2020, 578, 149–153. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).