A Proof-of-Concept Protein Microarray-Based Approach for Serotyping of Salmonella enterica Strains
Abstract
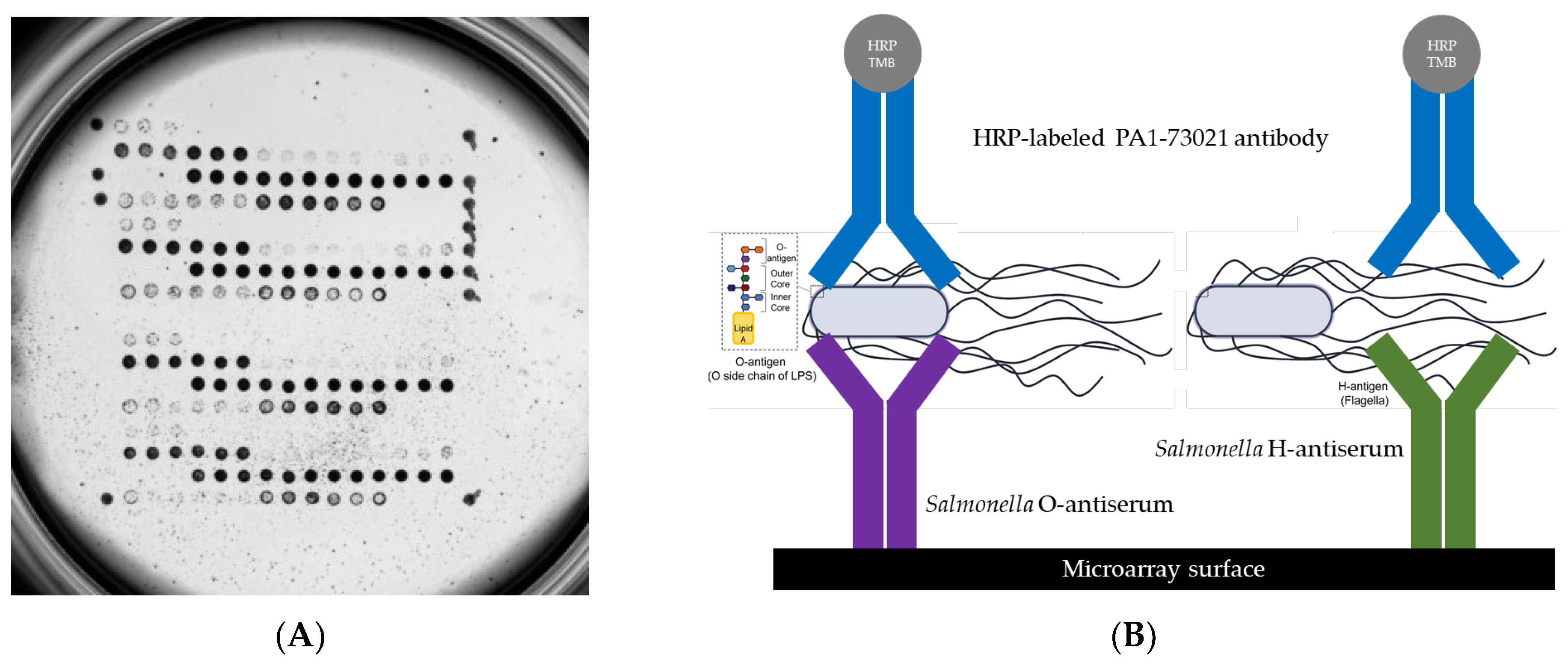
1. Introduction
2. Results and Discussion
3. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galan-Relano, A.; Valero Diaz, A.; Huerta Lorenzo, B.; Gomez-Gascon, L.; Mena Rodriguez, M.F.; Carrasco Jimenez, E.; Perez Rodriguez, F.; Astorga Marquez, R.J. Salmonella and salmonellosis: An update on public health implications and control strategies. Animals 2023, 13, 3666. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Salmonella infection—Prevention and treatment by antibiotics and probiotic yeasts: A review. Microbiology 2018, 164, 1327–1344. [Google Scholar] [CrossRef]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jove, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial Resistance Genes, Cassettes, and Plasmids Present in Salmonella enterica Associated With United States Food Animals. Front. Microbiol. 2019, 10, 832. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO: Geneva, Switzerland, 2007; Available online: https://www.pasteur.fr/sites/default/files/veng_0.pdf (accessed on 10 January 2024).
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.X. Supplement 2008–2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef]
- Barbour, E.K.; Ayyash, D.B.; Alturkistni, W.; Alyahiby, A.; Yaghmoor, S.; Iyer, A.; Yousef, J.; Kumosani, T.; Harakeh, S. Impact of sporadic reporting of poultry Salmonella serovars from selected developing countries. J. Infect. Dev. Ctries. 2015, 9, 1–7. [Google Scholar] [CrossRef]
- Mukherjee, N.; Nolan, V.G.; Dunn, J.R.; Banerjee, P. Sources of human infection by Salmonella enterica serotype Javiana: A systematic review. PLoS ONE 2019, 14, e0222108. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Wattiau, P.; Boland, C.; Bertrand, S. Methodologies for Salmonella enterica subsp. enterica subtyping: Gold standards and alternatives. Appl. Environ. Microbiol. 2011, 77, 7877–7885. [Google Scholar] [CrossRef]
- Fang, Z.; Wu, W.; Lu, X.; Zeng, L. Lateral flow biosensor for DNA extraction-free detection of Salmonella based on aptamer mediated strand displacement amplification. Biosens. Bioelectron. 2014, 56, 192–197. [Google Scholar] [CrossRef]
- Halcon, M.; Santos, J.A.P.; Lantican, N.B. Detection of Salmonella enterica subsp. enterica via quenching of unincorporated amplification signal reporters in loop-mediated isothermal amplification. Sci. World J. 2022, 2022, 4567817. [Google Scholar] [CrossRef]
- Wang, J.; Guo, K.; Li, S.; Liu, D.; Chu, X.; Wang, Y.; Guo, W.; Du, C.; Wang, X.; Hu, Z. Development and application of real-time PCR assay for detection of Salmonella Abortusequi. J. Clin. Microbiol. 2023, 61, e0137522. [Google Scholar] [CrossRef]
- Braun, S.D.; Ziegler, A.; Methner, U.; Slickers, P.; Keiling, S.; Monecke, S.; Ehricht, R. Fast DNA serotyping and antimicrobial resistance gene determination of salmonella enterica with an oligonucleotide microarray-based assay. PLoS ONE 2012, 7, e46489. [Google Scholar] [CrossRef]
- Banerji, S.; Simon, S.; Tille, A.; Fruth, A.; Flieger, A. Genome-based serotyping as the new gold standard. Sci. Rep. 2020, 10, 4333. [Google Scholar] [CrossRef]
- Zhang, X.M.; Payne, M.; Lan, R.T. Identification of serovar-specific genes for serotyping. Front. Microbiol. 2019, 10, 835. [Google Scholar] [CrossRef]
- Diep, B.; Barretto, C.; Portmann, A.C.; Fournier, C.; Karczmarek, A.; Voets, G.; Li, S.; Deng, X.; Klijn, A. Salmonella serotyping; comparison of the traditional method to a microarray-based method and an in silico platform using whole genome sequencing data. Front. Microbiol. 2019, 10, 2554. [Google Scholar] [CrossRef]
- Ibrahim, G.M.; Morin, P.M. Salmonella serotyping using whole genome sequencing. Front. Microbiol. 2018, 9, 2993. [Google Scholar] [CrossRef]
- Benjamin, W.H., Jr.; Posey, B.S.; Briles, D.E. Effects of in vitro growth phase on the pathogenesis of Salmonella typhimurium in mice. J. Gen. Microbiol. 1986, 132, 1283–1295. [Google Scholar] [CrossRef]
- Seligmann, E.; Wassermann, M.; Saphra, I. A new Salmonella type, Salmonella cubana. J. Bacteriol. 1946, 51, 123. [Google Scholar] [CrossRef]
- Popoff, M.Y.; Bockemuhl, J.; Gheesling, L.L. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 2004, 155, 568–570. [Google Scholar] [CrossRef]
- Litrup, E.; Torpdahl, M.; Malorny, B.; Huehn, S.; Helms, M.; Christensen, H.; Nielsen, E.M. DNA microarray analysis of Salmonella serotype Typhimurium strains causing different symptoms of disease. BMC Microbiol. 2010, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Chui, L.; Ferrato, C.; Li, V.; Christianson, S. Comparison of molecular and in silico Salmonella serotyping for Salmonella surveillance. Microorganisms 2021, 9, 955. [Google Scholar] [CrossRef]
- Cai, H.Y.; Lu, L.; Muckle, C.A.; Prescott, J.F.; Chen, S. Development of a novel protein microarray method for serotyping Salmonella enterica strains. J. Clin. Microbiol. 2005, 43, 3427–3430. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, specificity, and predictive values: Foundations, pliabilities, and pitfalls in research and practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
| Characteristics of Salmonella enterica Serovars, Salmonella bongori, and Controls | Antisera | TR1101_A-67_Vi | TR1111_A-E_Vi | TR1302_O:4 | TR1303_O:5 | TR1307_O:9 | TR1406_H1:g | TR1410_H1:i | TS1413_H1:m | TR1437_H2:1 | TR1433_H2:2 | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calculation of Concordance | ||||||||||||||||||||||||||||||||||||||||
| Salmonella enterica Subsp. enterica serovars and Controls | Somatic (O) Antigen | Flagellar Antigen H1 | Flagellar Antigen H2 | Number of Antisera | FN | FN in % | FP | FP in % | True (TP/TN) | Concordance | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison | Expected | Actual | Comparison |
| Abony | 1,4,[5],12,[27] | b | e,n,x | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Agama | 4,12 | i | 1,6 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true | P | P | true | N | N | true |
| Blegdam | 9,12 | g,m,q | - | 10 | 3 | 30% | 0 | 0% | 7 | 70% | P | N | FN | P | N | FN | N | N | true | N | N | true | P | P | true | P | P | true | N | N | true | P | N | FN | N | N | true | N | N | true |
| Brandenburg | 4,[5],12 | l,v | e,n,z15 | 10 | 0 | 0% | 1 | 10% | 9 | 90% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | P | FP | N | N | true |
| Bredeney | 1,4,12,27 | l,v | 1,7 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Breukelen | 6,8 | l,z13,[z28] | e,n,z15 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Budapest | 1,4,12,[27] | g,t | - | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true |
| California | 4,12 | g,m,t | [z67] | 10 | 1 | 10% | 1 | 10% | 8 | 80% | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | P | P | true | N | N | true | P | N | FN | N | P | FP | N | N | true |
| Choleraesuis | 6,7 | c | 1,5 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Corvallis | 8,20 | z4,z23 | [z6] | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Cubana | 1,13,23 | z29 | - | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Dublin | 1,9,12[Vi] | g,p | - | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Enteritidis | 1,9,12 | g,m | - | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | P | true | N | N | true | N | N | true | P | P | true | P | P | true | N | N | true | P | N | FN | N | N | true | N | N | true |
| Franken | 9,12 | z6 | z67 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Gallinarum | 1,9,12 | - | - | 10 | 1 | 10% | 0 | 0% | 9 | 90% | P | N | FN | P | P | true | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Gloucester | 1,4,12,27 | i | l,w | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true |
| Goeteborg | 9,12 | c | 1,5 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Heidelberg | 1,4,[5],12 | r | 1,2 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | P | P | true |
| Inverness | 38 | k | 1,6 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Kambole | 6,7 | d | 1,[2],7 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | P | N | FN |
| Mississippi | 1,13,23 | b | 1,5 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Montevideo | 6,7,14 | g,m,[p],s | [1,2,7] | 10 | 5 | 50% | 0 | 0% | 5 | 50% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true | P | N | FN | P | N | FN | P | N | FN |
| Moscow | 1,9,12 | g,q | - | 10 | 3 | 30% | 0 | 0% | 7 | 70% | P | N | FN | P | N | FN | N | N | true | N | N | true | P | N | FN | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Nitra | 2,12 | g,m | - | 10 | 3 | 30% | 0 | 0% | 7 | 70% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true | P | N | FN | N | N | true | N | N | true |
| Panama | 1,9,12 | l,v | 1,5 | 10 | 1 | 10% | 0 | 0% | 9 | 90% | P | P | true | P | N | FN | N | N | true | N | N | true | P | P | true | N | N | true | N | N | true | N | N | true | P | P | true | N | N | true |
| Paratyphi B | 1,4,[5],12 | b | 1,2 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | P | true | P | P | true | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | P | P | true |
| Potsdam | 6,7,14 | l,v | e,n,z15 | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Saintpaul | 1,4,[5],12 | e,h | 1,2 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | P | P | true |
| Singapore | 6,7 | k | e,n,x | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Stanleyville | 1,4,[5],12,[27] | z4,z23 | [1,2] | 10 | 1 | 10% | 0 | 0% | 9 | 90% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | N | N | true | N | N | true | P | P | true | P | N | FN |
| Typhimurium | 1,4,[5],12 | i | 1,2 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | P | P | true | P | P | true | P | P | true | P | P | true | N | N | true | N | N | true | P | P | true | N | N | true | P | P | true | P | P | true |
| Uno | 6,8 | z29 | [e,n,z15] | 10 | 2 | 20% | 0 | 0% | 8 | 80% | P | N | FN | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| S. bongori | 66 | z41 | - | 10 | 1 | 10% | 0 | 0% | 9 | 90% | P | N | FN | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Escherichia coli | 6 | - | 1 | 10 | 0 | 0% | 0 | 0% | 10 | 100% | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| negative control | - | - | - | 10 | 0 | 0% | 0 | 0% | 10 | 100% | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true | N | N | true |
| Overall Concordance | 86% | |||||||||||||||||||||||||||||||||||||||
| Antisera | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| O:A-67 and Vi | 42.86% | 39.39% | 100.00% | 100.00% | 9.09% |
| O:A-E and Vi | 54.29% | 50.00% | 100.00% | 100.00% | 15.79% |
| O:4 | 100.00% | 100.00% | 100.00% | 100.00% | 100.00% |
| O:5 | 97.14% | 85.71% | 100.00% | 100.00% | 96.55% |
| O:9 | 97.14% | 87.50% | 100.00% | 100.00% | 96.43% |
| H1:g | 100.00% | 100.00% | 100.00% | 100.00% | 100.00% |
| H1:i | 100.00% | 100.00% | 100.00% | 100.00% | 100.00% |
| H1:m | 85.71% | 0.00% | 100.00% | 0.00% | 85.71% |
| H2:1 | 91.43% | 92.86% | 90.48% | 86.67% | 95.00% |
| H2:2 | 91.43% | 57.14% | 100.00% | 100.00% | 90.32% |
| Overall | 86.00% | 71.26% | 99.05% | 88.67% | 78.89% |
| Order Number | Antiserum | Spotted Dilutions | |
|---|---|---|---|
| 1 | TR1101 | Anti-Salmonella A-67 + Vi | 1:6, 1:8, 1:10, 1:20 |
| 2 | TR1111 | Anti-Salmonella I (A-E + Vi) | 1:6, 1:8, 1:10, 1:20 |
| 3 | TR1307 | Anti-Salmonella O:9 | 1:6, 1:8, 1:10, 1:20 |
| 4 | TR1302 | Anti-Salmonella O:4 | 1:6, 1:8, 1:10, 1:20 |
| 5 | TR1303 | Anti-Salmonella O:5 | 1:6, 1:8, 1:10, 1:20 |
| 6 | TR1437 | Anti-Salmonella H2:1 | 1:6, 1:8, 1:10, 1:20 |
| 7 | TR1433 | Anti-Salmonella H2:2 | 1:6, 1:8, 1:10, 1:20 |
| 8 | TR1413 | Anti-Salmonella H1:m | 1:6, 1:8, 1:10, 1:20 |
| 9 | TR1410 | Anti-Salmonella H1:i | 1:6, 1:8, 1:10, 1:20 |
| 10 | TR1406 | Anti-Salmonella H1:g | 1:6, 1:8, 1:10, 1:20 |
| Number | Name | Subspecies | Somatic (O) Antigen | Flagellar Antigen H1 | Flagellar Antigen H2 |
|---|---|---|---|---|---|
| 1 | Abony | I | 1,4,[5],12,[27] | b | e,n,x |
| 2 | Agama | I | 4,12 | i | 1,6 |
| 3 | Blegdam | I | 9,12 | g,m,q | - |
| 4 | Brandenburg | I | 4,[5],12 | l,v | e,n,z15 |
| 5 | Bredeney | I | 1,4,12,27 | l,v | 1,7 |
| 6 | Breukelen | I | 6,8 | l,z13,[z28] | e,n,z15 |
| 7 | Budapest | I | 1,4,12,[27] | g,t | - |
| 8 | California | I | 4,12 | g,m,t | [z67] |
| 9 | Choleraesuis | I | 6,7 | c | 1,5 |
| 10 | Corvallis | I | 8,20 | z4,z23 | [z6] |
| 11 | Cubana | I | 1,13,23 | z29 | - |
| 12 | Dublin | I | 1,9,12[Vi] | g,p | - |
| 13 | Enteritidis | I | 1,9,12 | g,m | - |
| 14 | Franken | I | 9,12 | z6 | z67 |
| 15 | Gallinarum | I | 1,9,12 | - | - |
| 16 | Gloucester | I | 1,4,12,27 | i | l,w |
| 17 | Goeteborg | I | 9,12 | c | 1,5 |
| 18 | Heidelberg | I | 1,4,[5],12 | r | 1,2 |
| 19 | Inverness | I | 38 | k | 1,6 |
| 20 | Kambole | I | 6,7 | d | 1,[2],7 |
| 21 | Mississippi | I | 1,13,23 | b | 1,5 |
| 22 | Montevideo | I | 6,7,14 | g,m,[p],s | [1,2,7] |
| 23 | Moscow | I | 1,9,12 | g,q | - |
| 24 | Nitra | I | 2,12 | g,m | - |
| 25 | Panama | I | 1,9,12 | l,v | 1,5 |
| 26 | Paratyphi B | I | 1,4,[5],12 | b | 1,2 |
| 27 | Potsdam | I | 6,7,14 | l,v | e,n,z15 |
| 28 | Saintpaul | I | 1,4,[5],12 | e,h | 1,2 |
| 29 | Singapore | I | 6,7 | k | e,n,x |
| 30 | Stanleyville | I | 1,4,[5],12,[27] | z4,z23 | [1,2] |
| 31 | Typhimurium | I | 1,4,[5],12 | i | 1,2 |
| 32 | Uno | I | 6,8 | z29 | [e,n,z15] |
| 33 | Salmonella bongori | V | 66 | z41 | - |
| 34 | Escherichia coli | - | 6 | - | 1 |
| 35 | PBS buffer | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braun, S.D.; Müller, E.; Frankenfeld, K.; Gary, D.; Monecke, S.; Ehricht, R. A Proof-of-Concept Protein Microarray-Based Approach for Serotyping of Salmonella enterica Strains. Pathogens 2024, 13, 355. https://doi.org/10.3390/pathogens13050355
Braun SD, Müller E, Frankenfeld K, Gary D, Monecke S, Ehricht R. A Proof-of-Concept Protein Microarray-Based Approach for Serotyping of Salmonella enterica Strains. Pathogens. 2024; 13(5):355. https://doi.org/10.3390/pathogens13050355
Chicago/Turabian StyleBraun, Sascha D., Elke Müller, Katrin Frankenfeld, Dominik Gary, Stefan Monecke, and Ralf Ehricht. 2024. "A Proof-of-Concept Protein Microarray-Based Approach for Serotyping of Salmonella enterica Strains" Pathogens 13, no. 5: 355. https://doi.org/10.3390/pathogens13050355
APA StyleBraun, S. D., Müller, E., Frankenfeld, K., Gary, D., Monecke, S., & Ehricht, R. (2024). A Proof-of-Concept Protein Microarray-Based Approach for Serotyping of Salmonella enterica Strains. Pathogens, 13(5), 355. https://doi.org/10.3390/pathogens13050355





