Descriptive Epidemiology of Pathogens Associated with Acute Respiratory Infection in a Community-Based Study of K–12 School Children (2015–2023)
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting
2.2. Participants
2.3. Definition of ARI
2.4. Recruitment
2.5. Data and Specimen Collection
2.6. Laboratory Assessment
2.7. Statistical Analysis
2.8. Human Protections
3. Results
3.1. Student Demographics and Illness Characteristics
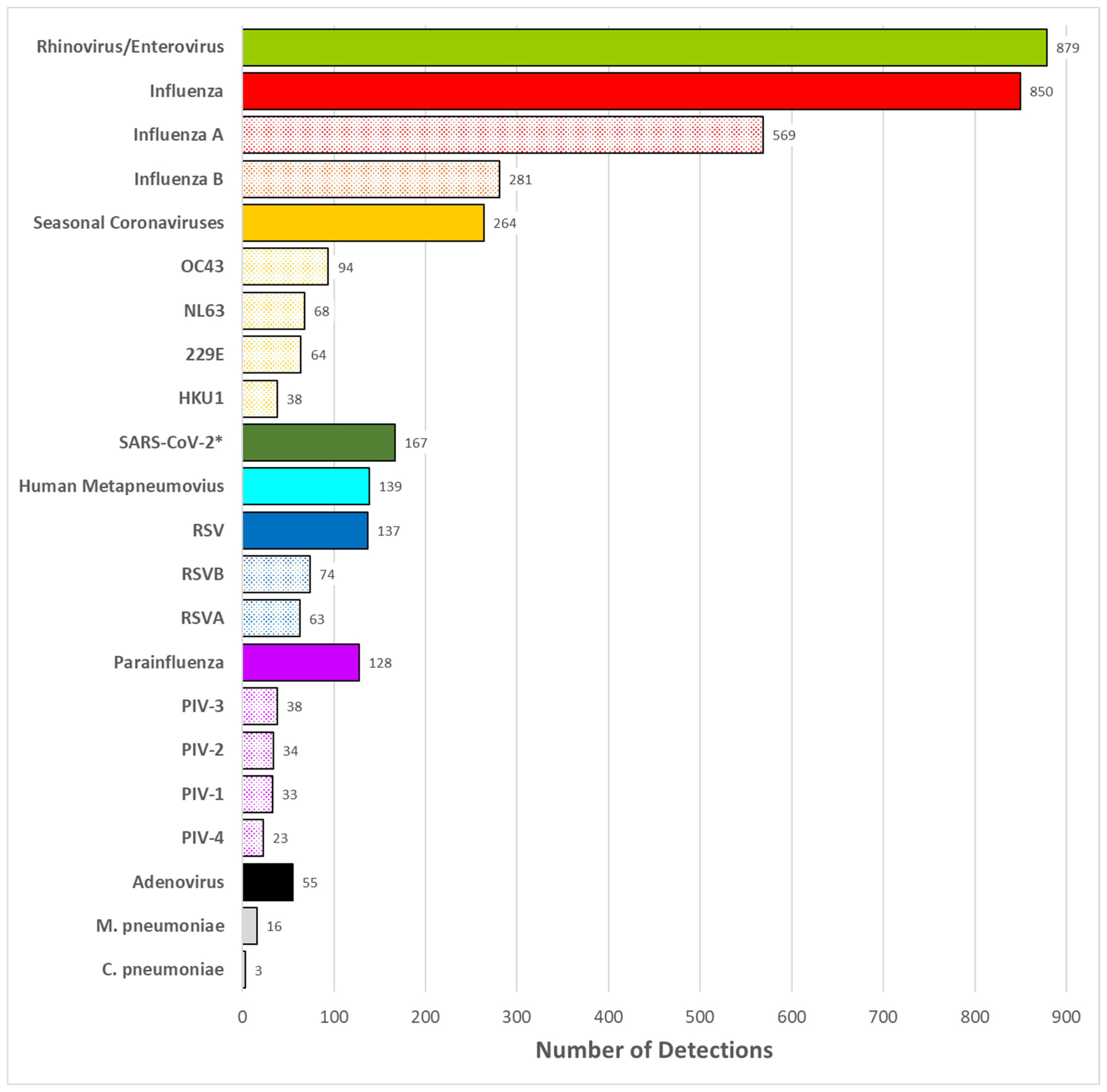
3.2. Pathogen Detection
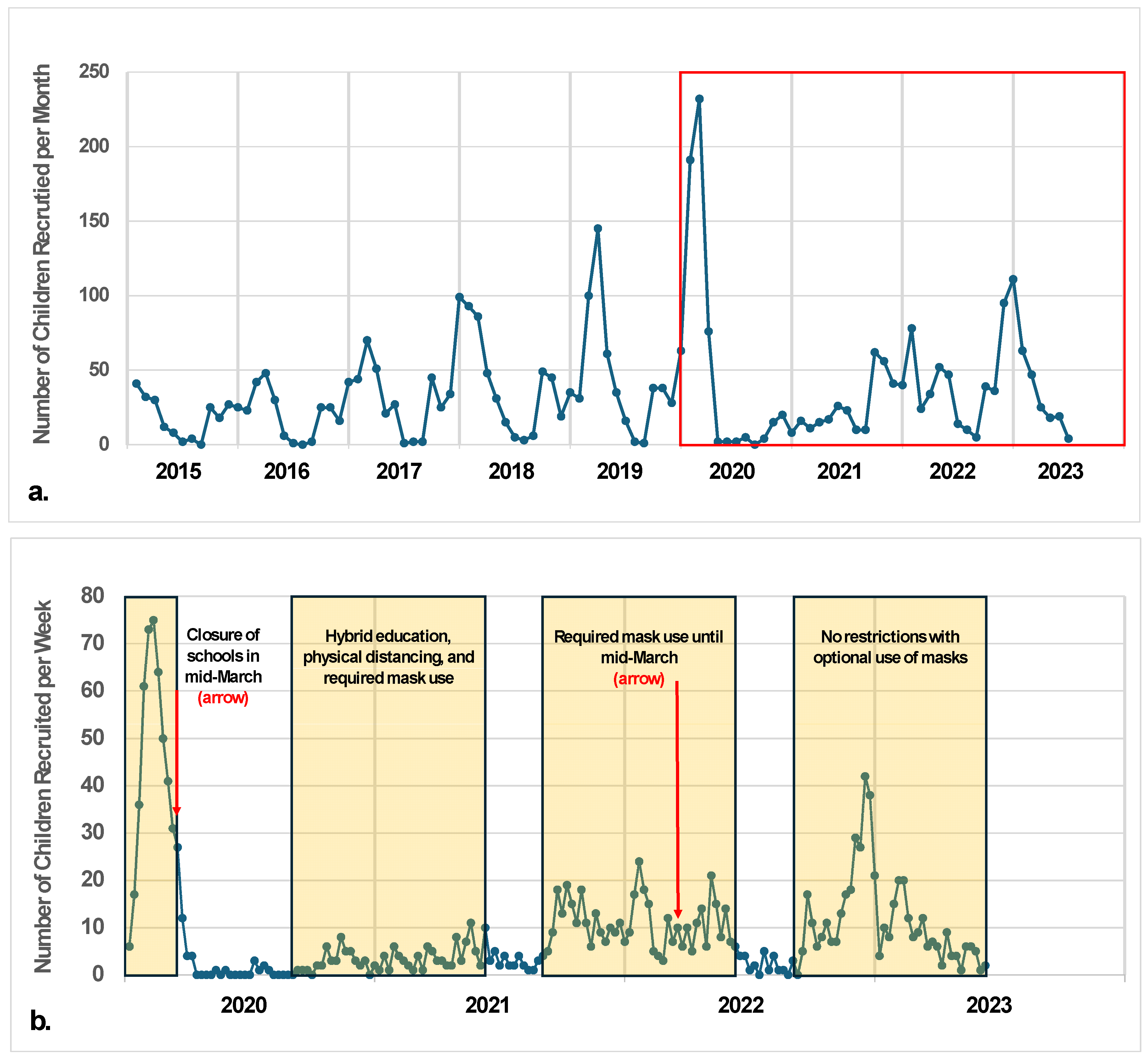
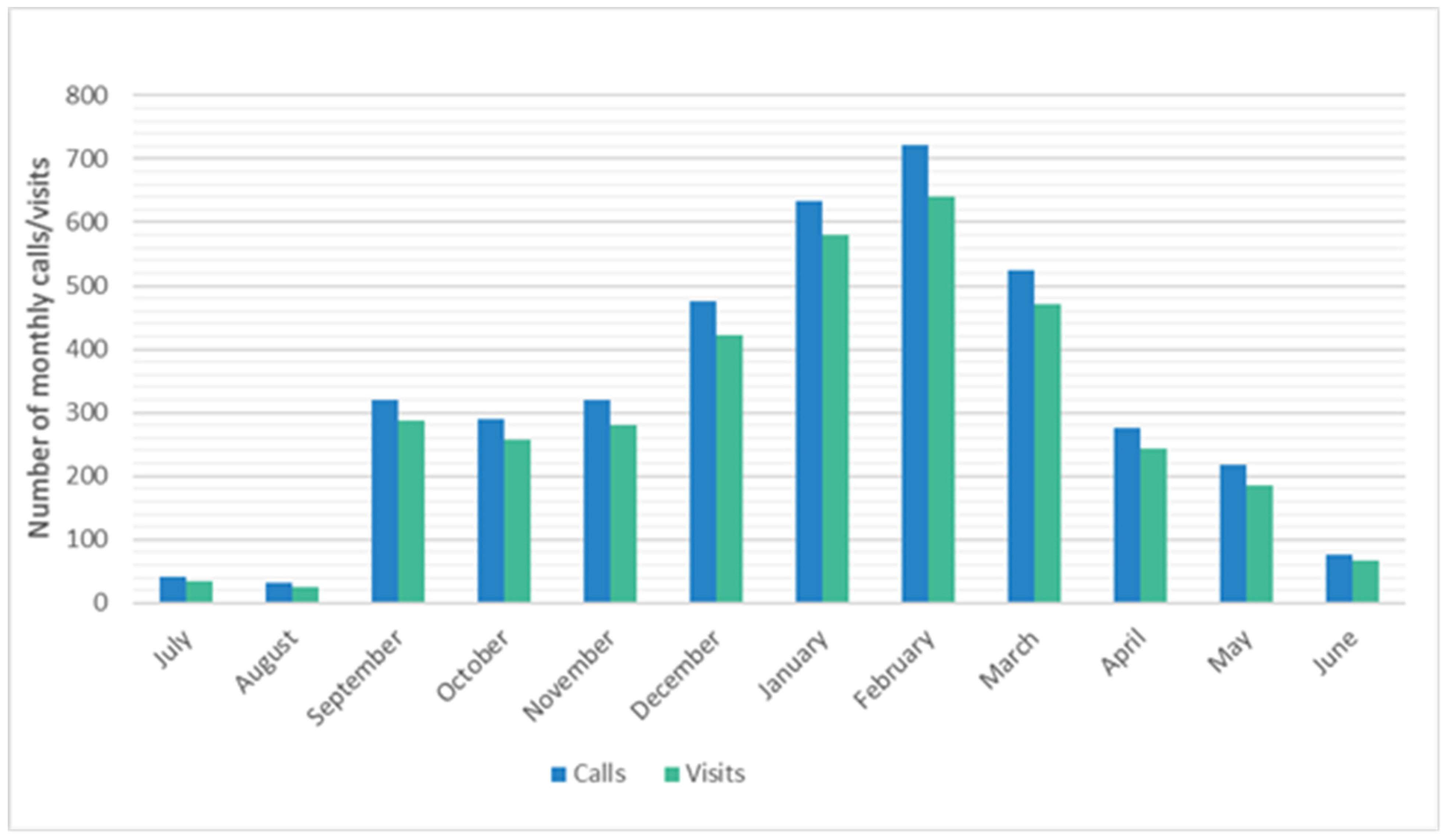
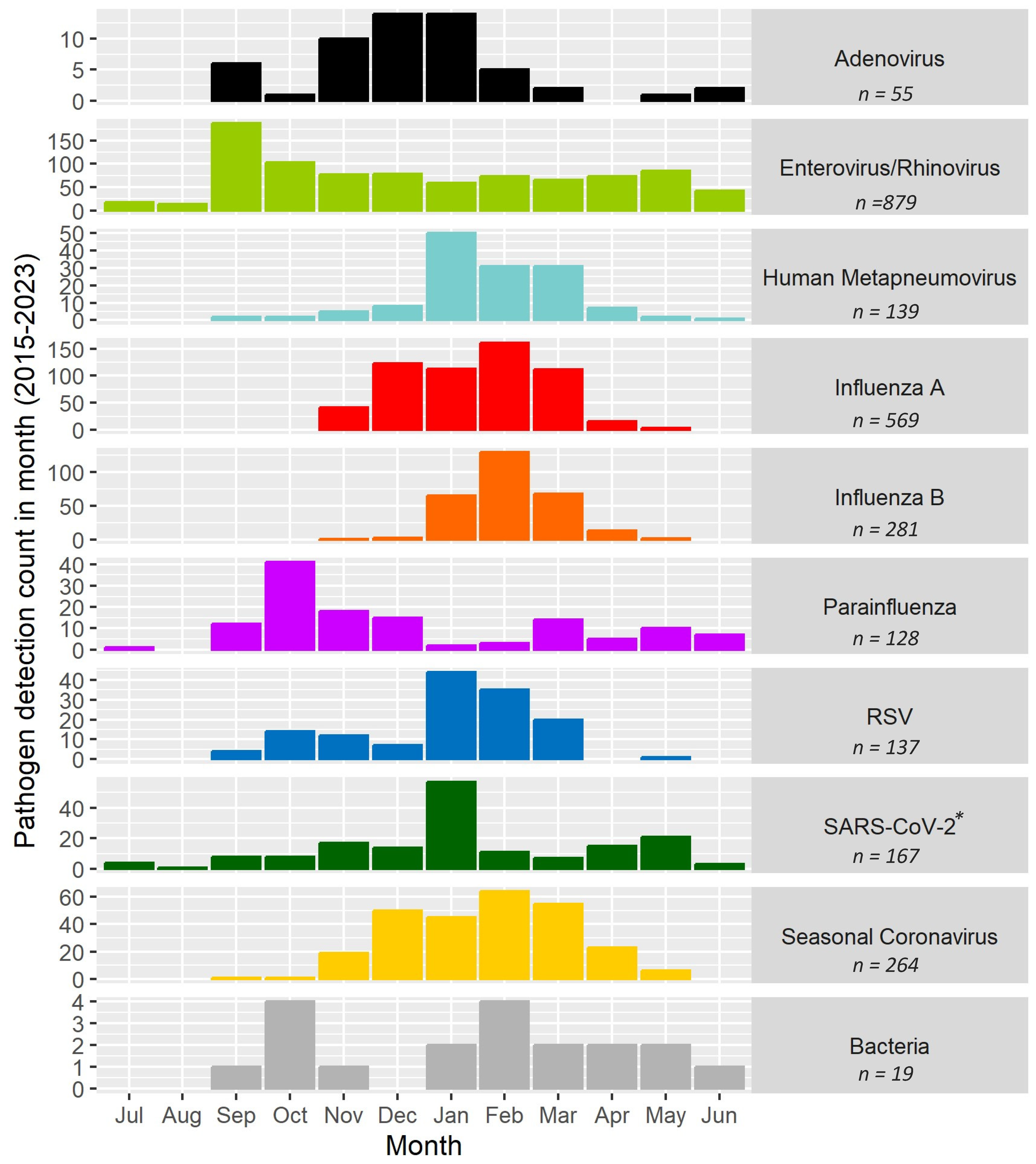
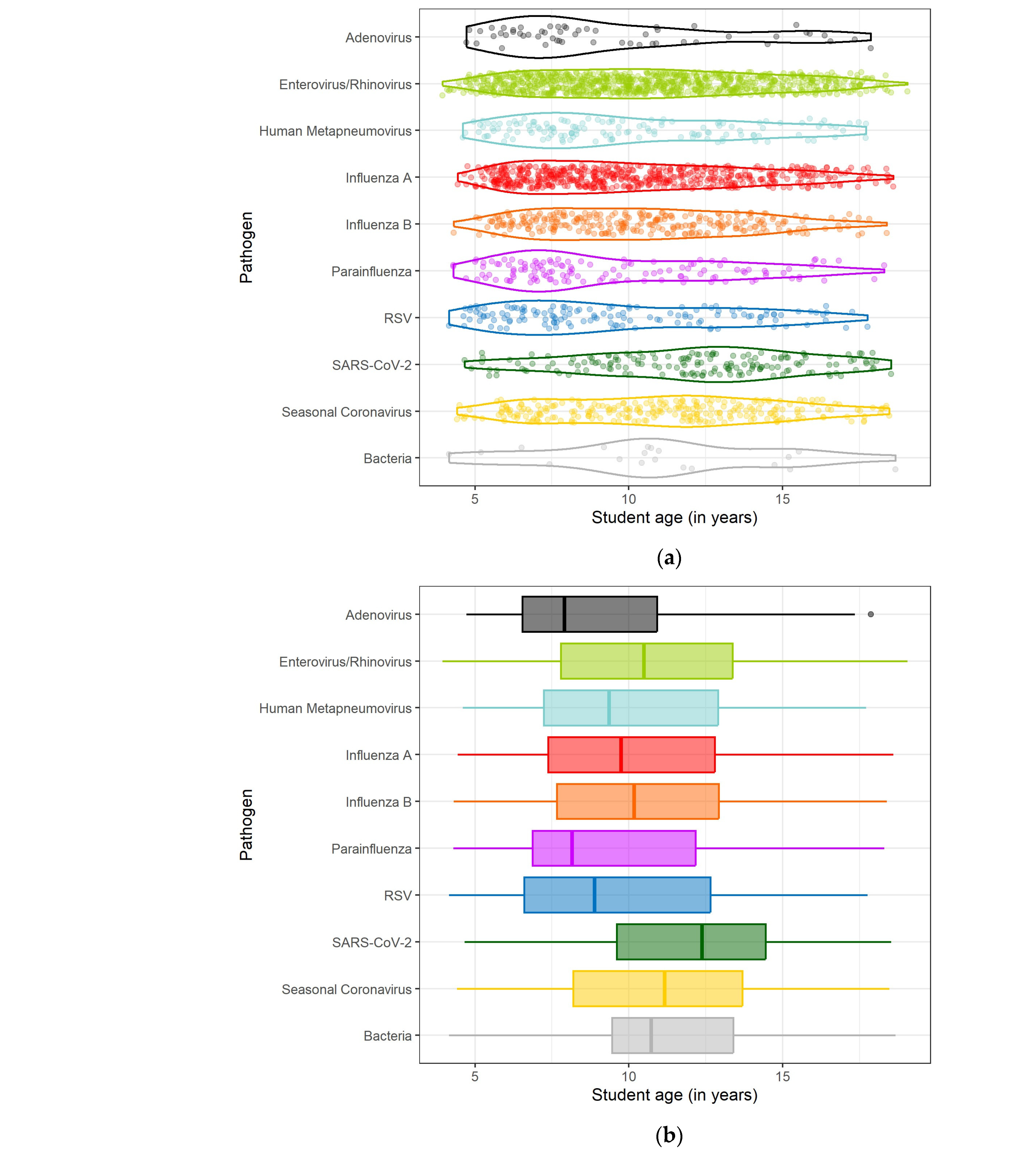
3.3. Timing and Seasonality
3.4. Self-Reported Symptoms and Illness Severity
4. Discussion
4.1. Limitations
4.2. Strengths/Lessons Learned
4.3. Summary of Findings and Implications
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monto, A.S.; Sullivan, K.M. Acute respiratory illness in the community. Frequency of illness and the agents involved. Epidemiol. Infect. 1993, 110, 145–160. [Google Scholar] [CrossRef]
- Temte, J.L.; Meiman, J.G.; Gangnon, R.E. School sessions are correlated with seasonal outbreaks of medically attended respiratory infections: Electronic health record time series analysis, Wisconsin 2004–2011. Epidemiol. Infect. 2019, 147, e127. [Google Scholar] [CrossRef] [PubMed]
- McLean, H.Q.; Peterson, S.H.; King, J.P.; Meece, J.K.; Belongia, E.A. School absenteeism among school-aged children with medically attended acute viral respiratory illness during three influenza seasons, 2012–2013 through 2014–2015. Influenza Other Respir. Viruses 2017, 11, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Bramley, T.J.; Lerner, D.; Sames, M. Productivity losses related to the common cold. J. Occup. Environ. Med. 2002, 44, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Temte, J.L.; Barlow, S.; Goss, M.; Temte, E.; Bell, C.; He, C.; Hamer, C.; Schemmel, A.; Maerz, B.; Comp, L.; et al. The Oregon child absenteeism due to respiratory disease study (ORCHARDS): Rationale, objectives, and design. Influenza Other Respir. Viruses 2022, 16, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, A.L.; Hardstaff, J.L.; Harris, J.P.; Vivancos, R.; O’brien, S.J. School-based surveillance of acute infectious disease in children: A systematic review. BMC Infect. Dis. 2021, 21, 744. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Cohen, A.L. Infectious Disease Surveillance. Int. Encycl. Public Health 2017, 222–229. [Google Scholar] [CrossRef] [PubMed Central]
- Widdowson, M.-A.; Monto, A.S. Epidemiology of influenza. In Textbook of Influenza, 2nd ed.; Webster, R.G., Monto, A.S., Braciale, T.J., Lamb, R.A., Eds.; Wiley Blackwell: Hoboken, NJ, USA, 2013; Chapter 16; p. 520. ISBN 978-0-470-67048-4. [Google Scholar]
- Chao, D.L.; Halloran, M.E.; Longini, I.M., Jr. School opening dates predict pandemic influenza A(H1N1) outbreaks in the United States. J. Infect. Dis. 2010, 202, 877–880. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gog, J.R.; Ballesteros, S.; Viboud, C.; Simonsen, L.; Bjornstad, O.N.; Shaman, J.; Chao, D.L.; Khan, F.; Grenfell, B.T. Spatial Transmission of 2009 Pandemic Influenza in the US. PLoS Comput. Biol. 2014, 10, e1003635. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Temte, J.L.; Barlow, S.; Goss, M.; Temte, E.; Schemmel, A.; Bell, C.; Reisdorf, E.; Shult, P.; Wedig, M.; Haupt, T.; et al. Cause-specific student absenteeism monitoring in K-12 schools for detection of increased influenza activity in the surrounding community—Dane County, Wisconsin, 2014–2020. PLoS ONE 2022, 17, e0267111. [Google Scholar] [CrossRef] [PubMed]
- Jackson, G.G.; Dowling, H.F.; Muldoon, R.L. Acute respiratory diseases of viral etiology. VII. Present concepts of the common cold. Am. J. Public Health Nations Health 1962, 52, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Al-Beltagi, M.; Saeed, N.K.; Bediwy, A.S.; El-Sawaf, Y. Paediatric gastrointestinal disorders in SARS-CoV-2 infection: Epidemiological and clinical implications. World J. Gastroenterol. 2021, 27, 1716–1727. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.T.; Temte, J.L.; Barlow, S.K.; Bell, C.J.; Goss, M.D.; Temte, E.G.; Checovich, M.M.; Reisdorf, E.; Scott, S.; Guenther, K.; et al. Comparison of participant-collected nasal and staff-collected oropharyngeal specimens for human ribonuclease P detection with RT-PCR during a community-based study. PLoS ONE 2020, 15, e0239000. [Google Scholar] [CrossRef] [PubMed]
- QuidelOrtho. Sofia Influenza A + B FIA. Available online: https://www.quidelortho.com/global/en/products/sofia-platform/sofia-influenza-a-b-fia (accessed on 20 December 2023).
- CDC. CDC Human Influenza Virus Real-Time RT-PCR Diagnostic Panel; Package Insert LB-029, R-0; CDC: Atlanta, GA, USA, 2011.
- ThermoFisher Scientific. TaqPath™ COVID-19, FluA, FluB Combo Kit Instructions for Use: 10 Multiplex Real-Time RT-PCR Test for the Detection and Differentiation of SARS-CoV-2, Influenza A, and Influenza B RNA. Available online: https://www.fda.gov/media/145931/download (accessed on 22 December 2023).
- Gonsalves, S.; Mahony, J.; Rao, A.; Dunbar, S.; Juretschko, S. Multiplexed detection and identification of respiratory pathogens using the NxTAG® respiratory pathogen panel. Methods 2019, 158, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zviedrite, N.; Hodis, J.D.; Jahan, F.; Gao, H.; Uzicanin, A. COVID-19-associated school closures and related efforts to sustain education and subsidized meal programs, United States, 18 February–30 June 2020. PLoS ONE 2021, 16, e0248925. [Google Scholar] [CrossRef]
- Temte, J.L.; Barlow, S.; Temte, E.; Goss, M.; Florek, K.; Braun, K.M.; Friedrich, T.C.; Reisdorf, E.; Bateman, A.C.; Uzicanin, A. Evidence of Early Household Transmission of SARS-CoV-2 Involving a School-aged Child. WMJ 2021, 120, 233–236. [Google Scholar]
- Goldman, J.L.; Lee, B.R.; Porter, J.; Deliu, A.; Tilsworth, S.; Almendares, O.M.; Sleweon, S.; Kirking, H.L.; Selvarangan, R.; Schuster, J.E. Notes from the Field: Multipathogen Respiratory Virus Testing Among Primary and Secondary School Students and Staff Members in a Large Metropolitan School District—Missouri, 2 November 2022–19 April 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 772–774. [Google Scholar] [CrossRef]
- Perez, A.; Lively, J.Y.; Curns, A.; Weinberg, G.A.; Halasa, N.B.; Staat, M.A.; Szilagyi, P.G.; Stewart, L.S.; McNeal, M.M.; Clopper, B.; et al. Respiratory Virus Surveillance Among Children with Acute Respiratory Illnesses—New Vaccine Surveillance Network, United States, 2016–2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 1253–1259. [Google Scholar] [CrossRef]
- Cash-Goldwasser, S.; Reingold, A.L.; Luby, S.P.; Jackson, L.A.; Frieden, T.R. Masks during Pandemics Caused by Respiratory Pathogens-Evidence and Implications for Action. JAMA Netw. Open 2023, 6, e2339443. [Google Scholar] [CrossRef] [PubMed]
- Temte, J.L.; Goss, M.; Bell, C.; Barlow, S.; Temte, E.; Bateman, A.; Uzicanin, A. Changing pattern of respiratory virus detections among school-aged children in a small community—Dane County, Wisconsin, September to December 2022. Influenza Other Respir. Viruses 2023, 17, e13171. [Google Scholar] [CrossRef] [PubMed]

| n (%) | |
|---|---|
| Total Population | 3498 |
| Age (mean, sd *) | 10.2 (3.52) |
| Male | 1957 (55.9) |
| White and non-Hispanic | 3175 (90.8) |
| Test Positivity (%) | |
| 2014–2015 | 82/129 (63.6) |
| 2015–2016 | 134/246 (54.5) |
| 2016–2017 | 190/324 (58.6) |
| 2017–2018 | 339/489 (69.3) |
| 2018–2019 | 373/539 (69.2) |
| 2019–2020 | 519/671 (77.4) |
| 2020–2021 | 123/175 (70.3) |
| 2021–2022 | 358/463 (77.3) |
| 2022–2023 | 352/465 (75.7) |
| Reported most likely source of infection | |
| Family member | 1070 (49.6) |
| School classmate | 747 (34.6) |
| Friend | 469 (21.7) |
| Self-reported severity of illness | |
| Mild | 843 (24.5) |
| Moderate | 2228 (64.7) |
| Severe | 373 (10.8) |
| Length of illness before home visit (mean, sd) | 2.29 (1.76) |
| Absent from school | 2633 (75.3) |
| Sought or planned to seek medical care | 421 (12.0) |
| Pathogen 1 | Pathogen 2 | Median Age Difference (Pathogen 1 Minus Pathogen 2) | Adjusted p Value |
|---|---|---|---|
| Adenovirus | Atypical Bacteria * | −2.81 | 0.0927 |
| Adenovirus | Enterovirus/Rhinovirus | −2.57 | 0.0068 |
| Adenovirus | Human Metapneumovirus | −1.45 | 0.2632 |
| Adenovirus | Influenza A | −1.83 | 0.0757 |
| Adenovirus | Influenza B | −2.26 | 0.0382 |
| Adenovirus | Parainfluenza | −0.24 | 0.6936 |
| Adenovirus | RSV ** | −0.97 | 0.5135 |
| Adenovirus | SARS-CoV-2 | −4.46 | 0.0000 |
| Adenovirus | Seasonal Coronavirus | −3.25 | 0.0013 |
| Atypical Bacteria * | Enterovirus/Rhinovirus | 0.24 | 0.7278 |
| Atypical Bacteria * | Human Metapneumovirus | 1.37 | 0.2952 |
| Atypical Bacteria * | Influenza A | 0.98 | 0.4085 |
| Atypical Bacteria * | Influenza B | 0.55 | 0.5383 |
| Atypical Bacteria * | Parainfluenza | 2.58 | 0.1223 |
| Atypical Bacteria * | RSV ** | 1.84 | 0.1654 |
| Atypical Bacteria * | SARS-CoV-2 | −1.65 | 0.2842 |
| Atypical Bacteria * | Seasonal Coronavirus | −0.44 | 0.9437 |
| Enterovirus/Rhinovirus | Human Metapneumovirus | 1.12 | 0.0461 |
| Enterovirus/Rhinovirus | Influenza A | 0.74 | 0.0260 |
| Enterovirus/Rhinovirus | Influenza B | 0.31 | 0.3715 |
| Enterovirus/Rhinovirus | Parainfluenza | 2.34 | 0.0008 |
| Enterovirus/Rhinovirus | RSV ** | 1.60 | 0.0029 |
| Enterovirus/Rhinovirus | SARS-CoV-2 | −1.89 | 0.0000 |
| Enterovirus/Rhinovirus | Seasonal Coronavirus | −0.68 | 0.1948 |
| Human Metapneumovirus | Influenza A | −0.38 | 0.5036 |
| Human Metapneumovirus | Influenza B | −0.81 | 0.2621 |
| Human Metapneumovirus | Parainfluenza | 1.21 | 0.3258 |
| Human Metapneumovirus | RSV ** | 0.48 | 0.4982 |
| Human Metapneumovirus | SARS-CoV-2 | −3.01 | 0.0000 |
| Human Metapneumovirus | Seasonal Coronavirus | −1.80 | 0.0070 |
| Influenza A | Influenza B | −0.43 | 0.4511 |
| Influenza A | Parainfluenza | 1.59 | 0.0562 |
| Influenza A | RSV ** | 0.86 | 0.1218 |
| Influenza A | SARS-CoV-2 | −2.63 | 0.0000 |
| Influenza A | Seasonal Coronavirus | −1.42 | 0.0034 |
| Influenza B | Parainfluenza | 2.02 | 0.0223 |
| Influenza B | RSV ** | 1.29 | 0.0507 |
| Influenza B | SARS-CoV-2 | −2.20 | 0.0000 |
| Influenza B | Seasonal Coronavirus | −0.99 | 0.0694 |
| Parainfluenza | RSV ** | −0.73 | 0.7297 |
| Parainfluenza | SARS-CoV-2 | −4.22 | 0.0000 |
| Parainfluenza | Seasonal Coronavirus | −3.01 | 0.0001 |
| RSV ** | SARS-CoV-2 | −3.49 | 0.0000 |
| RSV ** | Seasonal Coronavirus | −2.28 | 0.0004 |
| SARS-CoV-2 | Seasonal Coronavirus | 1.21 | 0.0089 |
| Self-Reported Symptoms (Proportions) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pathogen | Detections (n) | Fever | Cough | Runny Nose | Sore Throat | Fatigue | Stuffy Nose | Headache |
| SARS-CoV-2 | 167 | 0.49 | 0.34 | 0.33 | 0.34 | 0.41 | 0.35 | 0.30 |
| Influenza A | 570 | 0.83 | 0.93 | 0.72 | 0.68 | 0.86 | 0.79 | 0.66 |
| Influenza B | 283 | 0.87 | 0.94 | 0.75 | 0.75 | 0.92 | 0.79 | 0.71 |
| Adenovirus | 55 | 0.80 | 0.76 | 0.64 | 0.80 | 0.84 | 0.78 | 0.78 |
| Coronavirus 229E | 64 | 0.44 | 0.75 | 0.86 | 0.67 | 0.84 | 0.91 | 0.48 |
| Coronavirus HKU1 | 38 | 0.39 | 0.74 | 0.89 | 0.66 | 0.71 | 0.84 | 0.58 |
| Coronavirus NL63 | 67 | 0.40 | 0.87 | 0.78 | 0.85 | 0.70 | 0.84 | 0.49 |
| Coronavirus OC43 | 94 | 0.49 | 0.79 | 0.85 | 0.76 | 0.78 | 0.90 | 0.54 |
| Human Metapneumovirus | 139 | 0.59 | 0.96 | 0.73 | 0.70 | 0.81 | 0.88 | 0.49 |
| Parainfluenza 1 | 33 | 0.79 | 0.97 | 0.64 | 0.85 | 0.85 | 0.64 | 0.58 |
| Parainfluenza 2 | 34 | 0.68 | 0.97 | 0.47 | 0.88 | 0.85 | 0.71 | 0.50 |
| Parainfluenza 3 | 38 | 0.47 | 0.95 | 0.68 | 0.76 | 0.76 | 0.74 | 0.42 |
| Parainfluenza 4 | 23 | 0.43 | 0.83 | 0.83 | 0.70 | 0.78 | 0.87 | 0.39 |
| Enterovirus Rhinovirus | 880 | 0.35 | 0.77 | 0.84 | 0.78 | 0.72 | 0.88 | 0.52 |
| RSV * A | 63 | 0.57 | 0.98 | 0.76 | 0.76 | 0.75 | 0.86 | 0.59 |
| RSV * B | 73 | 0.52 | 0.97 | 0.81 | 0.71 | 0.79 | 0.92 | 0.59 |
| C. pneumoniae | 3 | 0.67 | 0.67 | 0.67 | 1.00 | 1.00 | 1.00 | 1.00 |
| M. pneumoniae | 16 | 0.81 | 1.00 | 0.38 | 0.63 | 0.88 | 0.44 | 0.69 |
| Totals | 2641 | 0.57 | 0.83 | 0.75 | 0.72 | 0.77 | 0.81 | 0.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bell, C.; Goss, M.; Norton, D.; Barlow, S.; Temte, E.; He, C.; Hamer, C.; Walters, S.; Sabry, A.; Johnson, K.; et al. Descriptive Epidemiology of Pathogens Associated with Acute Respiratory Infection in a Community-Based Study of K–12 School Children (2015–2023). Pathogens 2024, 13, 340. https://doi.org/10.3390/pathogens13040340
Bell C, Goss M, Norton D, Barlow S, Temte E, He C, Hamer C, Walters S, Sabry A, Johnson K, et al. Descriptive Epidemiology of Pathogens Associated with Acute Respiratory Infection in a Community-Based Study of K–12 School Children (2015–2023). Pathogens. 2024; 13(4):340. https://doi.org/10.3390/pathogens13040340
Chicago/Turabian StyleBell, Cristalyne, Maureen Goss, Derek Norton, Shari Barlow, Emily Temte, Cecilia He, Caroline Hamer, Sarah Walters, Alea Sabry, Kelly Johnson, and et al. 2024. "Descriptive Epidemiology of Pathogens Associated with Acute Respiratory Infection in a Community-Based Study of K–12 School Children (2015–2023)" Pathogens 13, no. 4: 340. https://doi.org/10.3390/pathogens13040340
APA StyleBell, C., Goss, M., Norton, D., Barlow, S., Temte, E., He, C., Hamer, C., Walters, S., Sabry, A., Johnson, K., Chen, G., Uzicanin, A., & Temte, J. (2024). Descriptive Epidemiology of Pathogens Associated with Acute Respiratory Infection in a Community-Based Study of K–12 School Children (2015–2023). Pathogens, 13(4), 340. https://doi.org/10.3390/pathogens13040340







