Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook
Abstract
1. Introduction
2. Taxonomy of Phages
3. Mechanisms of Infections
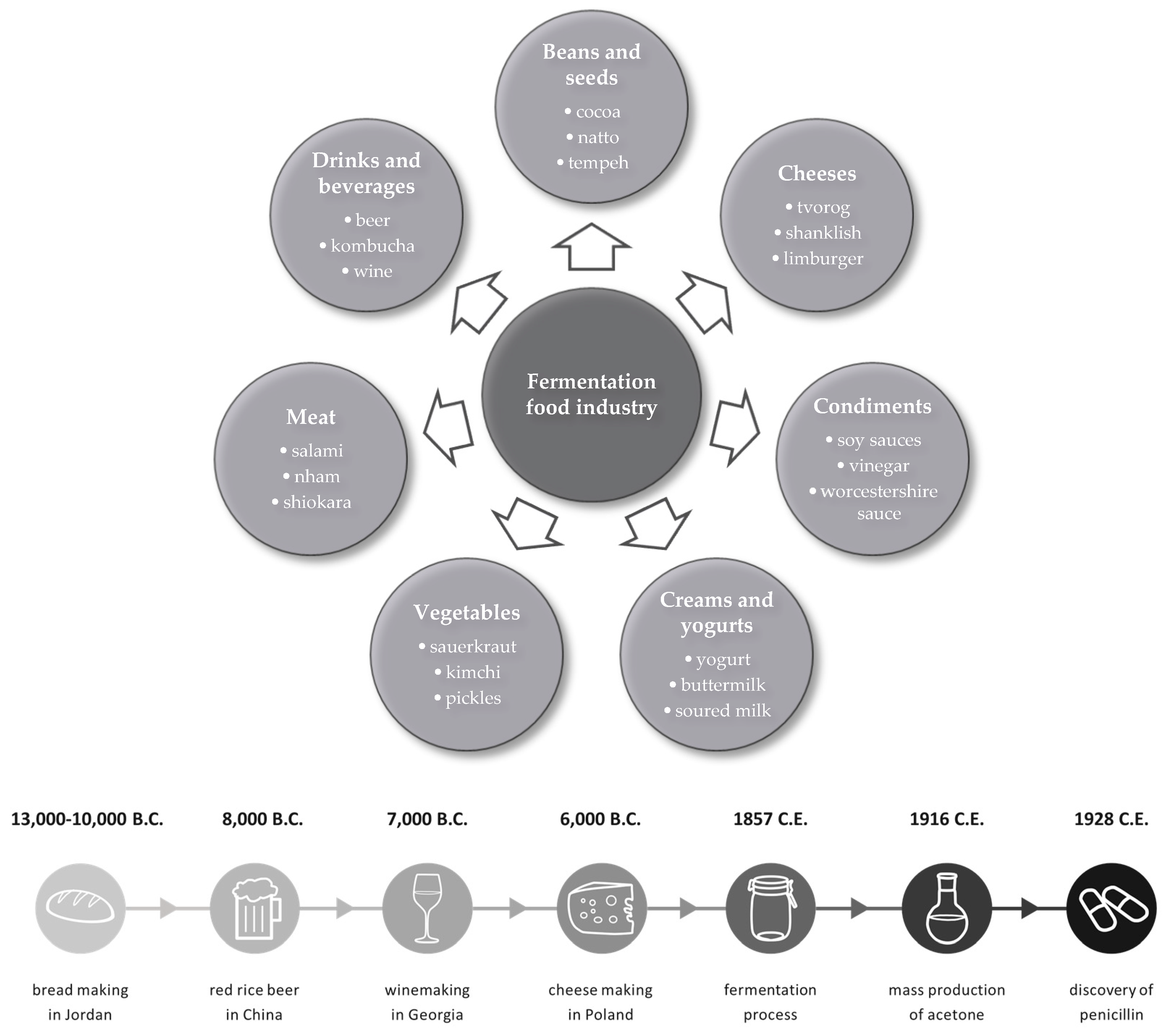
4. Fermentation Industry
5. Phage Infections
5.1. 1920s
5.2. 1930s
5.3. 1940s
5.4. 1950s
5.5. 1960s
5.6. 1970s
5.7. 1980s
5.8. 1990s
5.9. 2000s
5.10. 2010s
6. Future Outlook: New Antiphagents
6.1. Physical Factors
6.2. Chemical Factors
6.3. Antiphagents
7. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breitbart, M.; Rohwer, F. Here a Virus, There a Virus, Everywhere the Same Virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Curtis Hendrickson, R.; et al. Recent Changes to Virus Taxonomy Ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2022, 167, 2429–2440. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Laurinavičius, S.; Käkelä, R.; Bamford, D.H.; Somerharju, P. The Origin of Phospholipids of the Enveloped Bacteriophage Phi6. Virology 2004, 326, 182–190. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages Examined in the Electron Microscope. Arch. Virol. 2006, 152, 227–243. [Google Scholar] [CrossRef]
- Leprince, A.; Mahillon, J. Phage Adsorption to Gram-Positive Bacteria. Viruses 2023, 15, 196. [Google Scholar] [CrossRef] [PubMed]
- Makky, S.; Dawoud, A.; Safwat, A.; Abdelsattar, A.S.; Rezk, N.; El-Shibiny, A. The Bacteriophage Decides Own Tracks: When They Are with or against the Bacteria. Curr. Res. Microb. Sci. 2021, 2, 100050. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, M.; Łoś, J.M.; Łoś, M. Efficiency of Induction of Shiga-Toxin Lambdoid Prophages in Escherichia Coli Due to Oxidative and Antibiotic Stress Depends on the Combination of Prophage and the Bacterial Strain. J. Appl. Genet. 2020, 61, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.L. Phages Tune in to Host Cell Quorum Sensing. Cell 2019, 176, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Shin, H.; Lee, J.H.; Park, C.J.; Paik, S.Y.; Ryu, S. Isolation and Genome Characterization of the Virulent Staphylococcus Aureus Bacteriophage SA97. Viruses 2015, 7, 5225–5242. [Google Scholar] [CrossRef] [PubMed]
- Rakonjac, J.; Russel, M.; Khanum, S.; Brooke, S.J.; Rajič, M. Filamentous Phage: Structure and Biology. Adv. Exp. Med. Biol. 2017, 1053, 1–20. [Google Scholar] [CrossRef]
- Łoś, M.; Wegrzyn, G. Pseudolysogeny. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2012; Volume 82, pp. 339–349. [Google Scholar]
- Arranz-Otaegui, A.; Carretero, L.G.; Ramsey, M.N.; Fuller, D.Q.; Richter, T. Archaeobotanical Evidence Reveals the Origins of Bread 14,400 Years Ago in Northeastern Jordan. Proc. Natl. Acad. Sci. USA 2018, 115, 7925–7930. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; Zhao, Y.; Chen, X.; Gu, W. Beyond Subsistence: Evidence for Red Rice Beer in 8000-Year Old Neolithic Burials, North China. J. Archaeol. Sci. Reports 2023, 51, 104168. [Google Scholar] [CrossRef]
- Prajapati, J.B.; Nair, B.M. The History of Fermented Foods. In Handbook of Fermented Functional Foods; Farnwarth, E.R., Ed.; CRC Press: Boca Raton, FL, USA, 2003; pp. 1–26. ISBN 9780203009727. [Google Scholar]
- McGovern, P.; Jalabadze, M.; Batiuk, S.; Callahan, M.P.; Smith, K.E.; Hall, G.R.; Kvavadze, E.; Maghradze, D.; Rusishvili, N.; Bouby, L.; et al. Early Neolithic Wine of Georgia in the South Caucasus. Proc. Natl. Acad. Sci. USA 2017, 114, E10309–E10318. [Google Scholar] [CrossRef] [PubMed]
- Salque, M.; Bogucki, P.I.; Pyzel, J.; Sobkowiak-Tabaka, I.; Grygiel, R.; Szmyt, M.; Evershed, R.P. Earliest Evidence for Cheese Making in the Sixth Millennium BC in Northern Europe. Nature 2013, 493, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Gal, J. The Discovery of Biological Enantioselectivity: Louis Pasteur and the Fermentation of Tartaric Acid, 1857—A Review and Analysis 150 Yr Later. Chirality 2008, 20, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Walton, M.T.; Martin, J.L. Production of Butanol-Acetone by Fermentation; Peppler, H.J., Perlman, D., Eds.; Academic Press, Inc.: Cambridge, MA, USA, 1979; Volume 1, ISBN 0125515014. [Google Scholar]
- Ligon, B.L. Penicillin: Its Discovery and Early Development. Semin. Pediatr. Infect. Dis. 2004, 15, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Pasteur, L. Mémoire Sur La Fermentation Appelée Lactique. Comptes Rendus Chim. 1857, 45, 913–916. [Google Scholar]
- Jones, D.T.; Shirley, M.; Wu, X.; Keis, S. Bacteriophage Infections in the Industrial Acetone Butanol (AB) Fermentation Process. J. Mol. Microbiol. Biotechnol. 2000, 2, 21–26. [Google Scholar] [PubMed]
- Katz, L.; Baltz, R.H. Natural Product Discovery: Past, Present, and Future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- D’Este, M.; Alvarado-Morales, M.; Angelidaki, I. Amino Acids Production Focusing on Fermentation Technologies—A Review. Biotechnol. Adv. 2018, 36, 14–25. [Google Scholar] [CrossRef]
- Shrestha, S.; Chio, C.; Khatiwada, J.R.; Kognou, A.L.M.; Qin, W. Optimization of Multiple Enzymes Production by Fermentation Using Lipid-Producing Bacillus Sp. Front. Microbiol. 2022, 13, 1049692. [Google Scholar] [CrossRef]
- Wang, J.; Lin, M.; Xu, M.; Yang, S.T. Anaerobic Fermentation for Production of Carboxylic Acids as Bulk Chemicals from Renewable Biomass. In Advances in Biochemical Engineering/Biotechnology; Springer Science+Business Media: Berlin, Germany, 2016; Volume 156, pp. 323–363. [Google Scholar]
- Baeshen, N.A.; Baeshen, M.N.; Sheikh, A.; Bora, R.S.; Morsi, M.; Ahmed, M.M.; Ramadan, H.A.I.; Saini, K.S.; Redwan, E.M.; Abdullah, S.; et al. Cell Factories for Insulin Production. Microb. Cell Fact. 2014, 13, 141. [Google Scholar] [CrossRef]
- Moineau, S.; Lévesque, C. Control of Bacteriophages in Industrial Fermentations. In Bacteriophages—Biology and Applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, FL, USA, 2004; ISBN 9780849313363. [Google Scholar]
- Gabriel, C.L. Butanol Fermentation Process. Ind. Eng. Chem. 1928, 20, 1063–1067. [Google Scholar] [CrossRef]
- Ross, D. The Acetone-Butanol Fermentation. Prog Ind Microbiol. 1961, 3, 71–90. [Google Scholar]
- McCoy, E.; McDaniel, L.E.; Sylvester, J.C. Bacteriophages in a Butyl Fermentation Plant. J. Bacteriol. 1944, 47, 433. [Google Scholar]
- Whitehead, H.R.; Cox, G.A. Bacteriophage Phenomena in Cultures of Lactic Streptococci. J. Dairy Res. 1936, 7, 55–62. [Google Scholar] [CrossRef]
- Dmitrieff, C. The Phenomenon of Dissociation and Spontaneous Lysis in a Culture of an Actinomyces. Zhurnal Microbiol. Epidemiol. Immunobiol. 1934, 13, 289. [Google Scholar]
- Dmitrieff, C.; Souteeff, G. Sur Les Phenomenes de Dissociation Des Observes Dans Le Cultures de l’Actinomyces Bovis Bostroem Essais d’application Des Filtrate de Cultures Lysees Au Traitement de l’actinomycose. Ann. Inst. Pasteur 1936, 56, 470–476. [Google Scholar]
- Wieringa, K.T.; Wiebols, G.L. De Aardappelshurft En de Heterolyse Der Schurfparasiet. Tijdschr. Plantenziekten 1936, 42, 235–240. [Google Scholar]
- Wiebols, G.L.W.; Wieringa, K.T. Bacteriophagie Een Algemeen Voorkommend Verschijnsel; Veenman en Zonen: Wageningen, The Netherlands, 1936; Volume 16. [Google Scholar]
- Krassilnikov, N.A. The Phenomenon of Autolysis in Actinomycetales. Microbiology 1938, 7, 708–720. [Google Scholar]
- Ogata, S.; Hongo, M. Bacteriophages of the Genus Clostridium. Adv. Appl. Microbiol. 1979, 25, 241–273. [Google Scholar] [CrossRef]
- Kinoshita, S.; Itagaki, S.; Okumura, T.; Terada, M.; Tomh, T. Studies on Bacteriophages for Cl. acetobutylicum. Nippon Nōgeikagaku Kaishi 1952, 26, 104–107. [Google Scholar] [CrossRef]
- Hongo, M.; Murata, A. Bacteriophages of Clostridium Saccharoperbutylacetonicum. I. Some Characteristics of the Twelve Phages Obtained from the Abnormally Fermented Broths. Agric. Biol. Chem. 1965, 29, 1135–1145. [Google Scholar] [CrossRef]
- Hongo, M.; Murata, A. Bacteriophages of Clostridium Saccharoperbutylacetonicum. VI. Further Characterization of HM-Phages. Agric. Biol. Chem. 1966, 30, 913–916. [Google Scholar] [CrossRef]
- Saudek, E.C.; Colingsworth, D.R. A Bacteriophage in the Streptomycin Fermentation. J. Bacteriol. 1947, 54, 41. [Google Scholar] [PubMed]
- Baltz, R.H. Bacteriophage-Resistant Industrial Fermentation Strains: From the Cradle to CRISPR/Cas9. J. Ind. Microbiol. Biotechnol. 2018, 45, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, H.B.; Nunheimer, T.D.; Lee, S.B. A Bacterial Virus for Actinomyces griseus. J Bacteriol. 1947, 54, 535–541. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and Phage-Derived Proteins—Application Approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef]
- Reilly, H.C.; Harris, D.A.; Waksman, S.A. An Actinophage for Streptomyces griseus. J. Bacteriol. 1947, 54, 451–466. [Google Scholar] [CrossRef]
- Jones, D.; Metzger, H.J.; Schatz, A.; Waksman, S.A. Control of Gram-Negative Bacteria in Experimental Animals by Streptomycin. Science 1944, 100, 105. [Google Scholar] [CrossRef]
- Katz, L.; Chen, Y.Y.; Gonzalez, R.; Peterson, T.C.; Zhao, H.; Baltz, R.H. Synthetic Biology Advances and Applications in the Biotechnology Industry: A Perspective. J. Ind. Microbiol. Biotechnol. 2018, 45, 449–461. [Google Scholar] [CrossRef]
- Koerber, W.L.; Greenspan, G.; Langlykke, A.F. Observations on the Multiplication of Phages Affecting Streptomyces Griseus. J. Bacteriol. 1950, 60, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Weindling, R.; Kapros, C. A Virulent Actinophage Destructive to S. Aureofaciens Cultures and Chlortetracycline Fermentations. Bact. Proc. Soc. Am. Bact. 1951, 48. [Google Scholar]
- Weindling, R.; Tresner, H.D.; Backus, E.J. The Host-Range of a Streptomyces Aureofaciens Actinophage. Nature 1961, 189, 603. [Google Scholar] [CrossRef]
- Carvajal, F. Phage Problems in the Streptomycin Fermentation. Mycologia 1953, 45, 209–234. [Google Scholar] [CrossRef]
- Hongo, M.; Harada, R.; Akahoshi, K.; Nagata, K.; Takahashi, S. Butanol Fermentation. XXXI Use of Clostridium Saccharoperbutylacetonicum on a Plant Scale. J. Agric. Chem. Soc. Jpn. 1965, 39, 247–251. [Google Scholar]
- Hongo, M.; Murata, A.; Harada, R.; Akahoshi, K.; Nagata, K.; Takahashi, S. Butanol Fermentation. XXXII. Abnormal Fermentations in Acetone-Butanol Production on a Plant Scale by Use of Clostridium saccharoperbutylacetonicum. J. Agric. Chem. Soc. Jpn. 1965, 39, 247–251. [Google Scholar]
- Saxena, A.; Kumari, R.; Mukherjee, U.; Singh, P.; Lal, R. Draft Genome Sequence of the Rifamycin Producer Amycolatopsis Rifamycinica DSM 46095. Genome Announc. 2014, 2, 10–1128. [Google Scholar] [CrossRef]
- Thiemann, J.E.; Hengeller, C.; Virgilio, A. Rifamycin. XXV: A Group of Actinophages Active on Streptomyces mediterranei. Nature 1962, 17, 1104–1105. [Google Scholar] [CrossRef]
- Whitman, P.A.; Marshall, R.T. Isolation of Psychrophilic Bacteriophage-Host Systems from Refrigerated Food Products. Appl. Microbiol. 1971, 22, 220–223. [Google Scholar] [CrossRef]
- Vikram, A.; Callahan, M.T.; Woolston, J.W.; Sharma, M.; Sulakvelidze, A. Phage Biocontrol for Reducing Bacterial Foodborne Pathogens in Produce and Other Foods. Curr. Opin. Biotechnol. 2022, 78, 102805. [Google Scholar] [CrossRef]
- Petricciani, J.C.; Chu, F.C.; Johnson, J.B.; Meyer, H.M. Bacteriophages in Live Virus Vaccines. Proc. Soc. Exp. Biol. Med. 1973, 144, 789–791. [Google Scholar] [CrossRef]
- Haselkorn, R.; Schichman, S.; Milstien, J.; Petriccian, J. Characteristics of Bacteriophage ΦV-1 Isolated from Live Virus Vaccines. Proc. Soc. Exp. Biol. Med. 1978, 158, 383–387. [Google Scholar] [CrossRef]
- Petricciani, J.; Sheets, R.; Griffiths, E.; Knezevic, I. Adventitious Agents in Viral Vaccines: Lessons Learned from 4 Case Studies. Biologicals 2014, 42, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Merril, C.R.; Friedman, T.B.; Attallah, F.M.; Geuer, M.R.; Krell, K.; Yarkin, R. Isolation of Bacteriophages from Commercial Sera. In Vitro 1972, 8, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Kolata, G.B. Phage in Live Virus Vaccines: Are They Harmful to People? Science 1975, 187, 522–523. [Google Scholar] [CrossRef] [PubMed]
- Parveen, S.; Bishai, W.R.; Murphy, J.R. Corynebacterium Diphtheriae: Diphtheria Toxin, the Tox Operon, and Its Regulation by Fe2 + Activation of Apo-DtxR. Microbiol. Spectr. 2019, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Merril, C.; Geier, M.; Petricciani, J. Bacterial Virus Gene Expression in Human Cells. Nature 1971, 233, 198–400. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.C.; Schechtman, L.M.; Ts’o, P.O.P.; Borenfreund, E.; Bendich, A. The Attachment and Penetration of T7 DNA/Phage in Syrian Hamster Embryonic Cells. Biochim. Biophys. Acta 1976, 435, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Casas, V.; Maloy, S. Role of Bacteriophage-Encoded Exotoxins in the Evolution of Bacterial Pathogens. Futur. Microbiol. 2011, 6, 1461–1473. [Google Scholar] [CrossRef]
- Gembara, K.; Dąbrowska, K. Phage-Specific Antibodies. Curr. Opin. Biotechnol. 2021, 68, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, J.; Schnabl, B. Phage Therapy: Targeting Intestinal Bacterial Microbiota for the Treatment of Liver Diseases. JHEP Rep. 2023, 5, 100909. [Google Scholar] [CrossRef] [PubMed]
- Young, M.J.; Hall, L.M.L.; Merabishvilli, M.; Pirnay, J.P.; Clark, J.R.; Jones, J.D. Phage Therapy for Diabetic Foot Infection: A Case Series. Clin. Ther. 2023, 45, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Guo, W.; Yu, X.; Guo, C.; Zhou, N.; Guo, X.; Huang, R.L.; Li, Q.; Zhu, Y. Engineered M13 Phage as a Novel Therapeutic Bionanomaterial for Clinical Applications: From Tissue Regeneration to Cancer Therapy. Mater. Today Bio 2023, 20, 100612. [Google Scholar] [CrossRef] [PubMed]
- Milstien, J.B.; Walker, J.R.; Petricciani, J.C. Bacteriophages in Live Virus Vaccines: Lack of Evidence for Effects on the Genome of Rhesus Monkeys. Science 1977, 197, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.C.; Johnson, H.B.; Orr, H.C.; Probst, P.G.; Petricciani, J.C.; Johnson, J.B.; Orr, H.C.; Probst, P.G.; Petricciani, J.C. Bacterial Virus Contamination of Fetal Bovine Sera. In Vitro 1973, 9, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, T.; Poulin, J.M.; Maret, R.; Pousaz, R. Isolation of a Bacteriophage of Leuconostoc Mesenteroides from Dairy Products. J. Appl. Bacteriol. 1978, 44, 159–161. [Google Scholar] [CrossRef]
- Lawrence, R.C.; Heap, H.A.; Limsowtin, G.; Jarvis, A.W. Cheddar Cheese Starters: Current Knowledge and Practices of Phage Characteristics and Strain Selection. J. Dairy Sci. 1978, 61, 1181–1191. [Google Scholar] [CrossRef]
- Shimizu Kadota, M.; Tsuchida, N. Physical Mapping of the Virion and the Prophage DNAs of a Temperate Lactobacillus Phage ΦQfg. J. Gen. Microbiol. 1984, 130, 423–430. [Google Scholar] [CrossRef]
- Davis, C.; Silveira, N.F.; Fleet, G.H.; Silveira, F.A.; Fleet, G.H. Occurrence and Properties of Bacteriophages of Leuconostoc Oenos in Australian Wines. Appl. Environ. Microbiol. 1985, 50, 872–876. [Google Scholar] [CrossRef]
- Cazelles, O.; Gnaegi, F. Enquête Sur l’importance Pratique Du Problème Des Bactériophages Dans Le Vin. Rev. Suisse Vitic. Arboric. Hortic 1982, 14, 267–270. [Google Scholar]
- Gnaegi, F.; Cazelles, O.; Sozzi, T.; D’Amico, N. Connaissances Sur Les Bactériophages de Leuconostoc Oenos et Progrès Dans La Maîtrise. Rev. Suisse Vitic. Arboric. Horticutlure 1984, 16, 59–65. [Google Scholar]
- Sozzi, T.; Gnaegi, F. The Problem of Bacteriophages in Wine. Adv. Vitic. oenology Econ. Gain. Proc. 1983, 391–402. [Google Scholar]
- Ebner, H.; Sellmer, S.; Follman, H. Acetic Acid. In Biotechnology Set; Rehm, J., Reed, G., Eds.; Wiley: Hoboken, NJ, USA, 2001; pp. 383–397. ISBN 9783527257621. [Google Scholar]
- Teuber, M.; Andresen, A.A.; Sievers, M. Bacteriophage Problems in Vinegar Fermentations. Biotechnol. Lett. 1987, 9, 37–38. [Google Scholar] [CrossRef]
- Bradley, D. The Isolation and Morphology of Some New Bacteriophages Specific for Bacillus and Acetobacter Species. J. Gen. Microbiol. 1965, 41, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Schocher, A.J.; Kuhn, H.; Schindler, B.; Palleroni, N.J.; Despreaux, C.W.; Boublik, M.; Miller, P.A. Acetobacter Bacteriophage A-1. Arch. Microbiol 1979, 121, 193–197. [Google Scholar] [CrossRef]
- Robakis, N.; Palleroni, N.; Despreaux, C.W.; Boublik, M.; Baker, C.; Churn, P.; Claus, C.W. Isolation and Characterization of Two Phages for Gluconobacter oxydans. J. Gen. Microbiol. 1985, 131, 2467–2473. [Google Scholar] [CrossRef][Green Version]
- Wünsche, L.; Fischer, H.; Kiesel, B. Lysogenie Und Lysogene Konversion Bei Methylotrophen Bakterien. Zeitshrift Für Allgemaine Mikrobiolgie 1982, 23, 81–94. [Google Scholar] [CrossRef]
- Stamm, W.W.; Kittelmann, M.; Follmann, H.; Trüper, H.G.; Triiper, H.G. The Occurrence of Bacteriophages in Spirit Vinegar Fermentation. Appl. Microbiol. Biotechnol. 1989, 30, 41–46. [Google Scholar] [CrossRef]
- Sellmer, S.; Sievers, M.; Teuber, M. Morphology, Virulence and Epidemiology of Bacteriophage Particles Isolated from Industrial Vinegar Fermentations. Syst. Appl. Microbiol. 1992, 15, 610–616. [Google Scholar] [CrossRef]
- Koptides, M.; Barak, I.; Sisova, M.; Baloghova, E.; Ugorcakova, J.; Timko, J. Characterization of Bacteriophage BFK20 from Brevibacterium flavum. J. Gen. Microbiol. 1992, 138, 1387–1391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moineau, S.; Borkaev, M.; Holler, B.J.; Walker, S.A.; Kondo, J.K.; Vedamuthu, E.R.; Vandenbergh, P.A. Isolation and Characterization of Lactococcal Bacteriophages from Cultured Buttermilk Plants in the United States. J. Dairy Sci. 1996, 79, 2104–2111. [Google Scholar] [CrossRef]
- Moineau, S.; Fortier, J.; Ackermann, H.W.; Pandian, S. Characterization of Lactococcal Bacteriophages from Quebec Cheese Plants. Can. J. Microbiol. 1992, 38, 875–882. [Google Scholar] [CrossRef]
- Josephsen, J.; Andersen, N.; Behrndt, H.; Brandsborg, E.; Christiansen, G.; Hansen, M.B.; Hansen, S.; Nielsen, E.W.; Vogensen, F.K. An Ecological Study of Lytic Bacteriophages of Lactococcus Lactis Subsp. Cremoris Isolated in a Cheese Plant over a Five Year Period. Int. Dairy J. 1994, 4, 123–140. [Google Scholar] [CrossRef]
- Jarvis, A.W. Relationships by DNA Homology between Lactococcal Phages 7-9, P335 and New Zealand Industrial Lactococcal Phages. Int. Dairy J. 1995, 5, 355–366. [Google Scholar] [CrossRef]
- Brüssow, H.; Bruttin, A.; Desiere, F.; Lucchini, S.; Foley, S. Molecular Ecology and Evolution of Streptococcus Thermophilus Bacteriophages—A Review. Virus Genes 1998, 16, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Gindreau, E.; Lonvaud-Funel, A. Molecular Analysis of the Region Encoding the Lytic System from Oenococcus Oeni Temperate Bacteriophage Φ1OMC. FEMS Microbiol. Lett. 1999, 171, 231–238. [Google Scholar] [CrossRef]
- Sun, W.; Zhao, F.; Zhao, S.; Qin, L.; Guo, J.; Sci, J.L.F. Effects of the Bacteriophages on 2-Keto-D-Gluconic Acid Fermentation in Industrial Process. Food Sci. 2005, 26, 36–41. [Google Scholar]
- Zeng, W.; Cai, W.; Liu, L.; Du, G.; Chen, J.; Zhou, J. Efficient Biosynthesis of 2-Keto-D-Gluconic Acid by Fed-Batch Culture of Metabolically Engineered Gluconobacter Japonicus. Synth. Syst. Biotechnol. 2019, 4, 134–141. [Google Scholar] [CrossRef]
- Suárez, V.B.; Quiberoni, A.; Binetti, A.G.; Reinheimer, J.A. Thermophilic Lactic Acid Bacteria Phages Isolated from Argentinian Dairy Industries. J. Food Prot. 2002, 65, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Madera, C.; Monjardín, C.; Suárez, J.E. Milk Contamination and Resistance to Processing Conditions Determine the Fate of Lactococcus Lactis Bacteriophages in Dairies. Appl. Environ. Microbiol. 2004, 70, 7365–7371. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W.; DuBow, M.S. Viruses of Prokaryotes: General Properties of Bacteriophages; CRC Press Inc.: Boca Raton, FL, USA, 1987; Volume 1, ISBN 2674298170. [Google Scholar]
- Ogata, S. Bacteriophage Contamination in Industrial Processes. Biotechnol. Bioeng. 1980, 22, 177–193. [Google Scholar] [CrossRef]
- Wünsche, L. Importance of Bacteriophages in Fermentation Processes. Acta Biotechnol. 1989, 9, 395–419. [Google Scholar] [CrossRef]
- Zhang, X.; Kong, J.; Qu, Y. Isolation and Characterization of a Lactobacillus Fermentum Temperate Bacteriophage from Chinese Yogurt. J. Appl. Microbiol. 2006, 101, 857–863. [Google Scholar] [CrossRef]
- Halter, M.C.; Zahn, J.A. Characterization of a Novel Lytic Bacteriophage from an Industrial Escherichia Coli Fermentation Process and Elimination of Virulence Using a Heterologous CRISPR–Cas9 System. J. Ind. Microbiol. Biotechnol. 2018, 45, 153–163. [Google Scholar] [CrossRef]
- Zahn, A.J.; Halter, C.M. Surveillance and Elimination of Bacteriophage Contamination in an Industrial Fermentation Process. In Bacteriophages—Perspectives and Future; IntechOpen: London, UK, 2018; pp. 1–18. [Google Scholar] [CrossRef][Green Version]
- Pringsulaka, O.; Patarasinpaiboon, N.; Suwannasai, N.; Atthakor, W.; Rangsiruji, A. Isolation and Characterisation of a Novel Podoviridae-Phage Infecting Weissella cibaria N 22 from Nham, a Thai Fermented Pork Sausage. Food Microbiol. 2011, 28, 518–525. [Google Scholar] [CrossRef]
- Marcó, M.B.; Moineau, S.; Quiberoni, A. Bacteriophages and Dairy Fermentations. Bacteriophage 2012, 2, 149–158. [Google Scholar] [CrossRef]
- Mahony, J.; Murphy, J.; Van Sinderen, D. Lactococcal 936-Type Phages and Dairy Fermentation Problems: From Detection to Evolution and Prevention. Front. Microbiol. 2012, 3, 335. [Google Scholar] [CrossRef] [PubMed]
- Pujato, S.A.; Quiberoni, A.; Mercanti, D.J. Bacteriophages on Dairy Foods. J. Appl. Microbiol. 2019, 126, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Łoś, M. Strategies of Phage Contamination Prevention in Industry. Open J. Bacteriol. 2020, 4, 20–23. [Google Scholar] [CrossRef]
- Sing, W.D.; Klaenhammer, T.R. A Strategy for Rotation of Different Bacteriophage Defenses in a Lactococcal Single-Strain Starter Culture System. Appl. Environ. Microbiol. 1993, 59, 365–372. [Google Scholar] [CrossRef]
- Raza, S.; Wdowiak, M.; Paczesny, J. An Overview of Diverse Strategies to Inactivate Enterobacteriaceae-Targeting Bacteriophages. EcoSal Plus 2023, 11, eesp00192022. [Google Scholar] [CrossRef]
- Marcó, M.B.; Suárez, V.B.; Quiberoni, A.; Pujato, S.A. Inactivation of Dairy Bacteriophages by Thermal and Chemical Treatments. Viruses 2019, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.G.; Behrens, M.A.; Streletzky, K.A.; Olsson, U.; Evilevitch, A. Portal Stability Controls Dynamics of DNA Ejection from Phage. J. Phys. Chem. 2016, 120, 6421–6429. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.W.; Evilevitch, A. Influence of Internal DNA Pressure on Stability and Infectivity of Phage λ. J. Mol. Biol. 2015, 427, 3189–3200. [Google Scholar] [CrossRef] [PubMed]
- Brié, A.; Bertrand, I.; Meo, M.; Boudaud, N.; Gantzer, C. The Effect of Heat on the Physicochemical Properties of Bacteriophage MS2. Food Environ. Virol. 2016, 8, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Guglielmotti, D.M.; Mercanti, D.J.; Reinheimer, J.A.; Quiberoni, A.d.L. Review: Efficiency of Physical and Chemical Treatments on the Inactivation of Dairy Bacteriophages. Front. Microbiol. 2012, 2, 282. [Google Scholar] [CrossRef]
- Ahmed, H.; Tamminen, L.M.; Emanuelson, U. Temperature, Productivity, and Heat Tolerance: Evidence from Swedish Dairy Production. Clim. Chang. 2022, 175, 1–18. [Google Scholar] [CrossRef]
- Bolumar, T.; Orlien, V.; Sikes, A.; Aganovic, K.; Bak, K.H.; Guyon, C.; Stübler, A.S.; de Lamballerie, M.; Hertel, C.; Brüggemann, D.A. High-Pressure Processing of Meat: Molecular Impacts and Industrial Applications. Compr. Rev. Food Sci. Food Saf. 2021, 20, 332–368. [Google Scholar] [CrossRef]
- Smiddy, M.; Kelly, A.L.; Patterson, M.F.; Hill, C. High Pressure-Induced Inactivation of Qβ Coliphage and C2 Phage in Oysters and in Culture Media. Int. J. Food Microbiol. 2006, 106, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Redepath, J.L.; Grossweiner, L.I. Radiation Inactivation of T7 Phage. Radiat. Res. 1978, 73, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Chang, P.H.; Hartert, J.; Wigginton, K.R. Reactivity of Enveloped Virus Genome, Proteins, and Lipids with Free Chlorine and UV254. Environ. Sci. Technol. 2018, 52, 7698–7708. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yanagisawa, F.; Fujita, H.; Ohkido, M. Inactivation of T4 Bacteriophage by near Ultraviolet Light in the Presence of 8-methoxypsoralen. J. Dermatol. 1978, 5, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Protein Isoelectric Point. In Bioinformatics and the Cell; Springer: Boston, MA, USA, 2007; pp. 207–219. ISBN 978-0-387-71336-6. [Google Scholar]
- Zandi, R.; Reguera, D. Mechanical Properties of Viral Capsids. Phys. Rev. E Stat. Nonlinear, Soft Matter Phys. 2005, 72, 021917. [Google Scholar] [CrossRef] [PubMed]
- Gelbart, W.M.; Knobler, C.M. The Physics of Phages. Phys. Today 2008, 61, 42–47. [Google Scholar] [CrossRef]
- Shima, S.; Fukuhara, Y.; Sakai, H. Inactivation of Bacteriophages by ε-Poly-L-Lysine Produced by Streptomyces. Agric. Biol. Chem. 1982, 46, 1917–1919. [Google Scholar] [CrossRef]
- Shalitin, C.; Danon, D.; Katchalski, E. Inactivation of Escherichia Coli Bacteriophage T2 by Poly-L-Lysine. I. Nature of the Inactivation Process. Arch. Biochem. Biophys. 1962, 99, 494–507. [Google Scholar] [CrossRef]
- Shalitin, C.; Katchalski, E. Inactivation of Escherichia Coli Bacteriophage T2 by Poly-L-Lysine. II. Properties of the Irreversibly Inactivated Phage. Arch. Biochem. Biophys. 1962, 99, 508–516. [Google Scholar] [CrossRef]
- Marton, H.L.; Kilbride, P.; Ahmad, A.; Sagona, A.P.; Gibson, M.I. Anionic Synthetic Polymers Prevent Bacteriophage Infection. J. Am. Chem. Soc. 2023, 145, 8794–8799. [Google Scholar] [CrossRef]
- Xiaowei, S.; Zivanovic, S.; D’Souza, D.H. Effect of Chitosan on the Infectivity of Murine Norovirus, Feline Calicivirus, and Bacteriophage MS2. J. Food Prot. 2009, 72, 2623–2628. [Google Scholar] [CrossRef]
- Jarach, N.; Dodiuk, H.; Kenig, S. Polymers in the Medical Antiviral Front-Line. Polymers 2020, 12, 1727. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Fan, M.; Wang, Z.; Wu, W.; Wang, J. Thermal and Chemical Inactivation of Lactobacillus Virulent Bacteriophage. J. Dairy Sci. 2017, 100, 7041–7050. [Google Scholar] [CrossRef]
- Agún, S.; Fernández, L.; González-Menéndez, E.; Martínez, B.; Rodríguez, A.; García, P. Study of the Interactions between Bacteriophage PhiIPLA-RODI and Four Chemical Disinfectants for the Elimination of Staphylococcus aureus Contamination. Viruses 2018, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Murphy, J.; Mahony, J.; Lugli, G.A.; Ventura, M.; Noben, J.P.; Franz, C.M.A.P.; Neve, H.; Nauta, A.; Van Sinderen, D. Biocidal Inactivation of Lactococcus Lactis Bacteriophages: Efficacy and Targets of Commonly Used Sanitizers. Front. Microbiol. 2017, 8, 107. [Google Scholar] [CrossRef]
- Branston, S.D.; Stanley, E.C.; Ward, J.M.; Keshavarz-Moore, E. Determination of the Survival of Bacteriophage M13 from Chemical and Physical Challenges to Assist in Its Sustainable Bioprocessing. Biotechnol. Bioprocess Eng. 2013, 18, 560–566. [Google Scholar] [CrossRef]
- Capra, M.L.; Quiberoni, A.; Reinheimer, J.A. Thermal and Chemical Resistance of Lactobacillus Casei and Lactobacillus Paracasei Bacteriophages. Lett. Appl. Microbiol. 2004, 38, 499–504. [Google Scholar] [CrossRef]
- Halfhide, D.E.; Gannon, B.W.; Hayes, C.M.; Roe, J.M. Wide Variation in Effectiveness of Laboratory Disinfectants against Bacteriophages. Lett. Appl. Microbiol. 2008, 47, 608–612. [Google Scholar] [CrossRef]
- De Siqueira, R.S.; Dodd, C.E.R.; Rees, C.E.D. Evaluation of the Natural Virucidal Activity of Teas for Use in the Phage Amplification Assay. Int. J. Food Microbiol. 2006, 111, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, W.; Saeed, F.; Ajaz, M.; Rasool, S.A. In Vitro Antimicrobial, Antibiofilm and Antiphage Activity of Thyme (Thymus Vulgaris). Pakistan J. Bot. 2022, 54, 1121–1128. [Google Scholar] [CrossRef]
- Ordon, M.; Nawrotek, P.; Stachurska, X.; Schmidt, A.; Mizielińska, M. Mixtures of Scutellaria Baicalensis and Glycyrrhiza L. Extracts as Antibacterial and Antiviral Agents in Active Coatings. Coatings 2021, 11, 1438. [Google Scholar] [CrossRef]
- Ordon, M.; Zdanowicz, M.; Nawrotek, P.; Stachurska, X.; Mizielińska, M. Polyethylene Films Containing Plant Extracts in the Polymer Matrix as Antibacterial and Antiviral Materials. Int. J. Mol. Sci. 2021, 22, 13438. [Google Scholar] [CrossRef]
- Adams, M.H. The Stability of Bacterial Viruses in Solutions of Salts. J. Gen. Physiol. 1949, 32, 579–594. [Google Scholar] [CrossRef]
- Beard, P.J. Decrease Resisntance of Purified Bacteriophage to Disinfectants. Sci. Proc. 1929, 26, 880–881. [Google Scholar] [CrossRef]
- Ahmad, A.; Farhan Asad, S.; Singh, S.; Hadi, S.M. DNA Breakage by Resveratrol and Cu(II): Reaction Mechanism and Bacteriophage Inactivation. Cancer Lett. 2000, 154, 29–37. [Google Scholar] [CrossRef]
- Kessler, D.; Krause, R.M. Inactivation of Streptococcal Bacteriophage by Sulfhydryl Reagents. Proc. Soc. Exp. Biol. Med. 1963, 114, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Molan, K.; Rahmani, R.; Krklec, D.; Brojan, M.; Stopar, D. Phi 6 Bacteriophage Inactivation by Metal Salts, Metal Powders, and Metal Surfaces. Fundam. Electrochem. Depos. 2006, 14, 25–40. [Google Scholar] [CrossRef]
- Fonstein, L.M.; Suraikina, T.I.; Tal, E.K.; Moshkovsky, Y.S. Effect of Palladium and Platinum Salts on Bacteriophage T4 and Its Isolated DNA. Genetika 1975, 11, 128–134. [Google Scholar]
- Shooter, K.V.; Howse, R.; Merrifield, R.K.; Robins, A.B. The Interaction of Platinum II Compounds with Bacteriophages T7 and R17. Chem. Biol. Interact. 1972, 5, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.; Folga, M.; Łoś, M.; Foltynowicz, Z.; Paczesny, J. The Effect of Zero-Valent Iron Nanoparticles (NZVI) on Bacteriophages. Viruses 2022, 14, 867. [Google Scholar] [CrossRef] [PubMed]
- Richter, Ł.; Paszkowska, K.; Cendrowska, U.; Olgiati, F.; Silva, P.J.; Gasbarri, M.; Guven, Z.P.; Paczesny, J.; Stellacci, F. Broad-Spectrum Nanoparticles against Bacteriophage Infections. Nanoscale 2021, 13, 18684–18694. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, X.; Polla, R.L.; Silva, P.J.; Ong, Q.K.; Stellacci, F. Ligand Concentration Determines Antiviral Efficacy of Silica Multivalent Nanoparticles. J. Colloid Interface Sci. 2024, 657, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Bonnain, C.; Breitbart, M.; Buck, K.N. The Ferrojan Horse Hypothesis: Iron-Virus Interactions in the Ocean. Front. Mar. Sci. 2016, 3, e99569. [Google Scholar] [CrossRef]
- Gutierrez, L.; Li, X.; Wang, J.; Nangmenyi, G.; Economy, J.; Kuhlenschmidt, T.B.; Kuhlenschmidt, M.S.; Nguyen, T.H. Adsorption of Rotavirus and Bacteriophage MS2 Using Glass Fiber Coated with Hematite Nanoparticles. Water Res. 2009, 43, 5198–5208. [Google Scholar] [CrossRef]
- Bradley, I.; Straub, A.; Maraccini, P.; Markazi, S.; Nguyen, T.H. Iron Oxide Amended Biosand Filters for Virus Removal. Water Res. 2011, 45, 4501–4510. [Google Scholar] [CrossRef]
- Briggiler Marcó, M.; Quiberoni, A.d.L.; Negro, A.C.; Reinheimer, J.A.; Alfano, O.M. Evaluation of the Photocatalytic Inactivation Efficiency of Dairy Bacteriophages. Chem. Eng. J. 2011, 172, 987–993. [Google Scholar] [CrossRef]
- Heffron, J.; McDermid, B.; Mayer, B.K. Bacteriophage Inactivation as a Function of Ferrous Iron Oxidation. Environ. Sci. Water Res. Technol. 2019, 5, 1309–1317. [Google Scholar] [CrossRef]

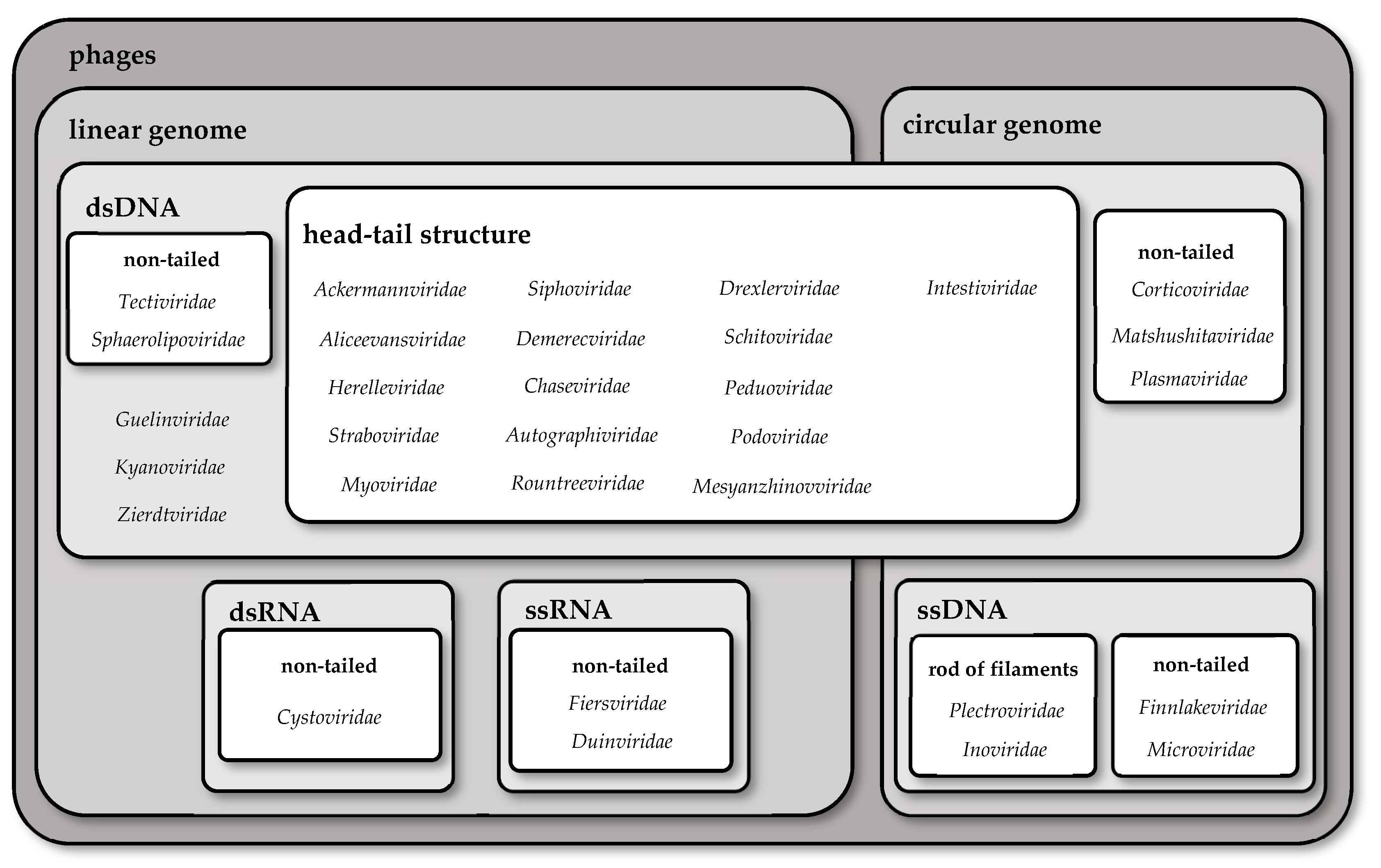
| Family | Shape of Genome | Nucleic Acid | Structure | Type of Tail |
|---|---|---|---|---|
| Ackermannviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Aliceevansviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Herelleviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Straboviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Myoviridae 1 | linear | dsDNA | head–tail structure | long, contractile tail |
| Mesyanzhinovviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Peduoviridae | linear | dsDNA | head–tail structure | long, contractile tail |
| Drexlerviridae | linear | dsDNA | head–tail structure | long, non-contractile tail |
| Siphoviridae 1 | linear | dsDNA | head–tail structure | long, non-contractile tail |
| Demerecviridae | linear | dsDNA | head–tail structure | long, non-contractile tail |
| Chaseviridae | linear | dsDNA | head–tail structure | long, non-contractile tail |
| Autographiviridae | linear | dsDNA | head–tail structure | short, non-contractile tail |
| Rountreeviridae | linear | dsDNA | head–tail structure | short, non-contractile tail |
| Podoviridae 1 | linear | dsDNA | head–tail structure | short, non-contractile tail |
| Schitoviridae | linear | dsDNA | head–tail structure | short, non-contractile tail |
| Tectiviridae | linear | dsDNA | non-tailed | - |
| Sphaerolipoviridae | linear | dsDNA | non-tailed | - |
| Guelinviridae | linear | dsDNA | - | - |
| Kyanoviridae | linear | dsDNA | - | - |
| Zierdtviridae | linear | dsDNA | - | - |
| Cystoviridae 2 | linear | dsRNA | non-tailed | - |
| Fiersviridae | linear | ssRNA (+) | non-tailed | - |
| Duinviridae | linear | ssRNA (+) | non-tailed | - |
| Intestiviridae | circular | dsDNA | head–tail structure | short, non-contractile tail |
| Corticoviridae | circular | dsDNA | non-tailed | - |
| Matshushitaviridae | circular | dsDNA | non-tailed | - |
| Plasmaviridae 2 | circular | dsDNA | non-tailed | - |
| Microviridae | circular | ssDNA | non-tailed | - |
| Finnlakeviridae | circular | ssDNA | non-tailed | - |
| Inoviridae | circular | ssDNA | rod of filaments | - |
| Plectroviridae | circular | ssDNA | rod of filaments | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamiński, B.; Paczesny, J. Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook. Pathogens 2024, 13, 152. https://doi.org/10.3390/pathogens13020152
Kamiński B, Paczesny J. Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook. Pathogens. 2024; 13(2):152. https://doi.org/10.3390/pathogens13020152
Chicago/Turabian StyleKamiński, Bartosz, and Jan Paczesny. 2024. "Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook" Pathogens 13, no. 2: 152. https://doi.org/10.3390/pathogens13020152
APA StyleKamiński, B., & Paczesny, J. (2024). Bacteriophage Challenges in Industrial Processes: A Historical Unveiling and Future Outlook. Pathogens, 13(2), 152. https://doi.org/10.3390/pathogens13020152






