Respiratory Pathogen Coinfection During Intersecting COVID-19 and Influenza Epidemics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Pathogen Testing
2.3. Data Visualization
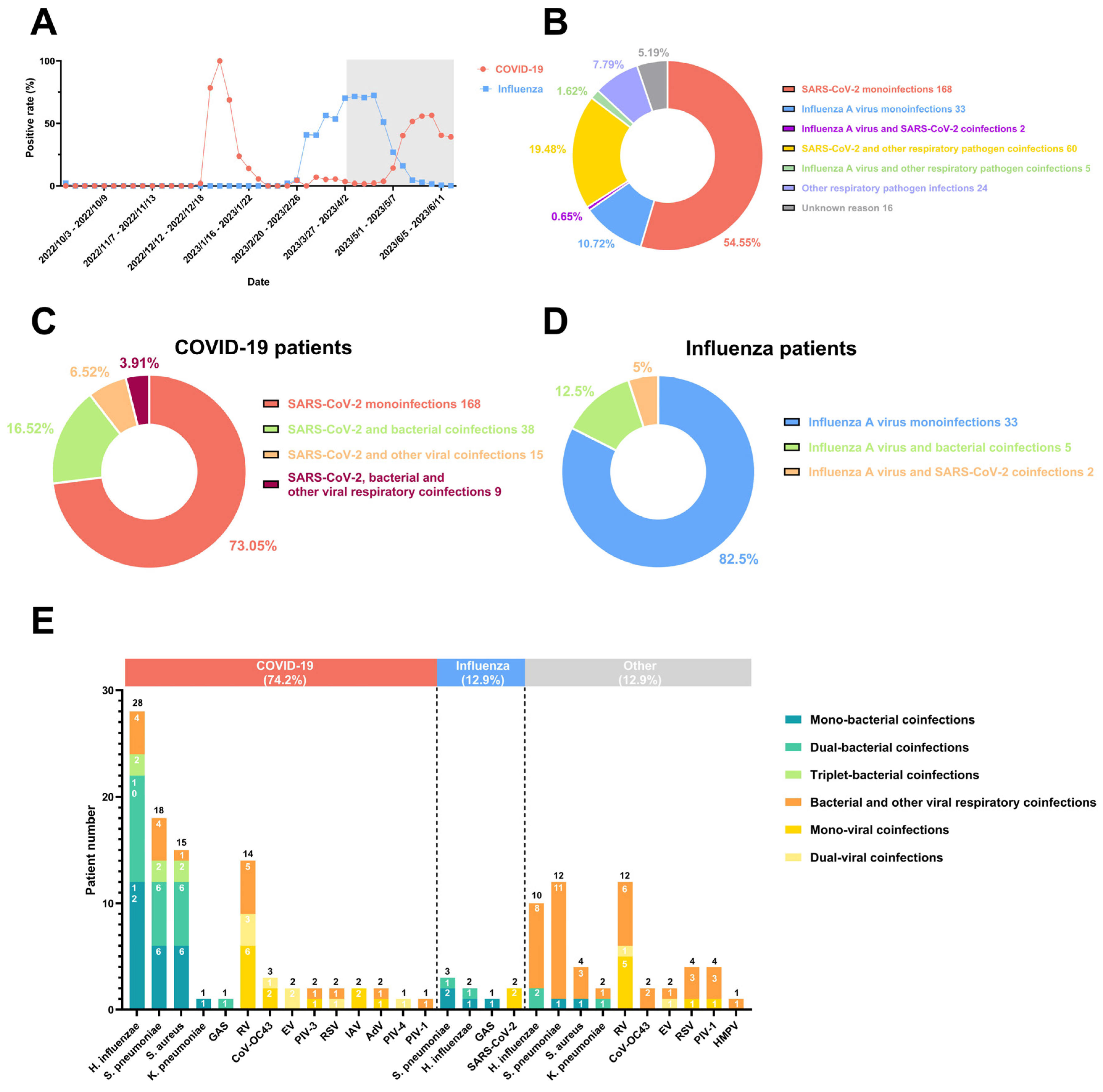
3. Results
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, G.H.; Belmonte, J.C. New Life is coming: Committed to improving human health. Life Med. 2022, 1, 1. [Google Scholar] [CrossRef]
- Westblade, L.F.; Simon, M.S.; Satlin, M.J. Bacterial Coinfections in Coronavirus Disease 2019. Trends Microbiol. 2021, 29, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Upadhyay, P.; Reddy, J.; Granger, J. SARS-CoV-2 respiratory co-infections: Incidence of viral and bacterial co-pathogens. Int. J. Infect. Dis. 2021, 105, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhou, L.; Lv, J.; Yang, S.; Chen, G.; Liu, X.; Han, C.; Tan, X.; Qian, S.; Wu, Z.; et al. Bacterial coinfections contribute to severe COVID-19 in winter. Cell Res. 2023, 33, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, H.; Su, Z.; Li, W.; Yang, D.; Deng, F.; Chen, J. Co-infection of SARS-CoV-2 and Influenza virus in Early Stage of the COVID-19 Epidemic in Wuhan, China. J. Infect. 2020, 81, e128–e129. [Google Scholar] [CrossRef] [PubMed]
- Swets, M.C.; Russell, C.D.; Harrison, E.M.; Docherty, A.B.; Lone, N.; Girvan, M.; Hardwick, H.E.; Visser, L.G.; Openshaw, P.J.; Groeneveld, G.H.; et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet 2022, 399, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Liu, Z.; Jiang, S. Double insult: Flu bug enhances SARS-CoV-2 infectivity. Cell Res. 2021, 31, 491–492. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Y.; Chang, P.-H.; Chen, K.-Y.; Lin, I.-F.; Hsih, W.-H.; Tsai, W.-L.; Chen, J.-A.; Lee, S.S.-J. Coronavirus disease 2019 (COVID-19) associated bacterial coinfection: Incidence, diagnosis and treatment. J. Microbiol. Immunol. Infect. 2022, 55, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Ito, M.; Furusawa, Y.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Adachi, E.; Saito, M.; Koga, M.; Tsutsumi, T.; Yamamoto, S.; et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect Dis. 2023, 23, 30–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Zheng, J.; Jin, Y.; Zhang, Y.; Tang, F.; Li, J.; Cheng, H.; Jiang, L.; Wen, H.; et al. Emerging Omicron subvariants evade neutralizing immunity elicited by vaccine or BA.1/BA.2 infection. J. Med. Virol. 2023, 95, e28539. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Jiang, L.; Chen, Q.; Fu, Y.; Jin, Y.; Chen, Z.; Tang, F.; Zeng, X.; Wen, H.; et al. Comparison of SARS-CoV-2 aerosol emission from patients with Omicron BA.1 or BA.2 subvariant infection. J. Infect. 2022, 85, e37–e39. [Google Scholar] [CrossRef]
- Jin, Y.; Cui, H.; Jiang, L.; Zhang, C.; Li, J.; Cheng, H.; Chen, Z.; Zheng, J.; Zhang, Y.; Fu, Y.; et al. Evidence for human infection with avian influenza A(H9N2) virus via environmental transmission inside live poultry market in Xiamen, China. J. Med. Virol. 2023, 95, e28242. [Google Scholar] [CrossRef]
- Li, J.; Zheng, J.; Chen, P.; Wang, B.; Zhang, Y.; Xiong, J.; You, L.; Jin, Y.; Jiang, L.; Tang, F.; et al. Higher SARS-CoV-2 shedding in exhaled aerosol probably contributed to the enhanced transmissibility of Omicron BA.5 subvariant. J. Med. Virol. 2023, 95, e28365. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; Pawelec, G. Single-cell immune atlas for human aging and frailty. Life Med. 2022, 1, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Mina, M.J.; Klugman, K.P. The role of influenza in the severity and transmission of respiratory bacterial disease. Lancet Respir. Med. 2014, 2, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Nickbakhsh, S.; Mair, C.; Matthews, L.; Reeve, R.; Johnson, P.C.D.; Thorburn, F.; von Wissmann, B.; Reynolds, A.; McMenamin, J.; Gunson, R.N.; et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA 2019, 116, 27142–27150. [Google Scholar] [CrossRef]
- Bai, L.; Zhao, Y.; Dong, J.; Liang, S.; Guo, M.; Liu, X.; Wang, X.; Huang, Z.; Sun, X.; Zhang, Z.; et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021, 31, 395–403. [Google Scholar] [CrossRef] [PubMed]
| COVID-Positive | Influenza-Positive | Other | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <18 | 18–44 | 45–64 | 65+ | <18 | 18–44 | 45–64 | 65+ | <18 | 18–44 | 45–64 | 65+ | |
| Total number, N | 42 (67.8%) | 16 (25.8%) | 3 (4.8%) | 1 (1.6%) | 2 (28.6) | 5 (71.4) | 0 | 0 | 13 (86.6%) | 1 (6.7%) | 1 (6.7%) | 0 |
| Testing | ||||||||||||
| SARS-CoV-2 | - | - | - | - | 0 | 2 (100%) | 0 | 0 | - | - | - | - |
| Influenza A virus | 0 | 2 (100%) | 0 | 0 | - | - | - | - | - | - | - | - |
| Haemophilus influenzae | 20 (71.4%) | 7 (25.0%) | 1 (3.6%) | 0 | 1 (50.0%) | 1 (50.0%) | 0 | 0 | 8 (100%) | 0 | 0 | 0 |
| Streptococcus pneumoniae | 15 (83.3%) | 2 (11.1%) | 0 | 1 (5.6%) | 2 (66.7%) | 1 (33.3%) | 0 | 0 | 7 (87.5%) | 1 (12.5%) | 0 | 0 |
| Rhinovirus | 11 (78.6%) | 3 (21.4%) | 0 | 0 | 0 | 0 | 0 | 0 | 6 (85.7%) | 0 | 1 (14.3%) | 0 |
| Staphylococcus aureus | 14 (93.3%) | 1 (6.7%) | 0 | 0 | 0 | 0 | 0 | 0 | 3 (100%) | 0 | 0 | 0 |
| Respiratory syncytial virus | 2 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (100%) | 0 | 0 | 0 |
| Coronavirus OC43 | 2 (66.7%) | 0 | 1 (33.3%) | 0 | 0 | 0 | 0 | 0 | 2 (100%) | 0 | 0 | 0 |
|
Parainfluenza virus
type I | 1 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (100%) | 0 | 0 | 0 |
| Enterovirus | 0 | 2 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (50.0%) | 0 | 1 (50.0%) | 0 |
|
Group A
Streptococcus | 1 (100%) | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 0 | 1 (100%) | 0 | 0 | 0 |
| Adenovirus | 2 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
Parainfluenza virus
type III | 1 (50.0%) | 0 | 1 (50.0%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
Klebsiella
pneumoniae | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
Human
metapneumovirus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Parainfluenza virus type IV | 1 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Jin, Y.; Li, J.; Zhang, R.; Zhang, Y.; Cheng, H.; Lu, B.; Zheng, J.; Li, L.; Wang, Z. Respiratory Pathogen Coinfection During Intersecting COVID-19 and Influenza Epidemics. Pathogens 2024, 13, 1113. https://doi.org/10.3390/pathogens13121113
Jiang L, Jin Y, Li J, Zhang R, Zhang Y, Cheng H, Lu B, Zheng J, Li L, Wang Z. Respiratory Pathogen Coinfection During Intersecting COVID-19 and Influenza Epidemics. Pathogens. 2024; 13(12):1113. https://doi.org/10.3390/pathogens13121113
Chicago/Turabian StyleJiang, Lina, Yifei Jin, Jingjing Li, Rongqiu Zhang, Yidun Zhang, Hongliang Cheng, Bing Lu, Jing Zheng, Li Li, and Zhongyi Wang. 2024. "Respiratory Pathogen Coinfection During Intersecting COVID-19 and Influenza Epidemics" Pathogens 13, no. 12: 1113. https://doi.org/10.3390/pathogens13121113
APA StyleJiang, L., Jin, Y., Li, J., Zhang, R., Zhang, Y., Cheng, H., Lu, B., Zheng, J., Li, L., & Wang, Z. (2024). Respiratory Pathogen Coinfection During Intersecting COVID-19 and Influenza Epidemics. Pathogens, 13(12), 1113. https://doi.org/10.3390/pathogens13121113








