Abstract
Pseudomonas aeruginosa is a leading cause of healthcare-associated infections, which are related to substantial morbidity and mortality. The incidence of Plasmid-Mediated Quinolone Resistance (PMQR) determinants has been previously reported in this bacterium. However, there is limited information regarding the presence of PMQR and carbapenemase-encoding genes simultaneously. This study aims to analyze the prevalence of these determinants on P. aeruginosa strain isolated from clinical patients in the State of Aguascalientes, Mexico. Fifty-two P. aeruginosa isolates from nosocomial patients were collected from Centenario Hospital Miguel Hidalgo. This is a retrospective observational study conducted at a single center. Antibiotic susceptibility was tested using the Vitek-2 system. Only carbapenem-resistant isolates were included in this study. Carbapenemase-encoding genes and PMQR determinants were screened by polymerase chain reaction (PCR). Resistance rates of 100% were found on tigecycline and ceftriaxone. Of the 52 isolates, 34.6% were positive for the qnr genes, 46.2% for the oqxA gene, and 25% for the aac-(6′)-lb gene. The most frequent carbapenemase genes found in the samples were blaOXA-51 (42.3%), blaOXA-1 (15.4%), and blaVIM (15.4%). blaOXA-51 co-carrying oqxA was detected in 21.1% of the isolates, blaOXA-51 co-carrying aac-(6’)-lb in 11.5%, blaVIM co-carrying aac-(6′)-lb in 3.8%, and blaKPC co-carrying oqxA in 5.8%. Systematic surveillance to detect carbapenemase-encoding genes and PMQR determinants, and rational prescription using the last-line drugs could help in preventing the dissemination of multidrug-resistant determinants.
1. Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium that causes significant healthcare-related illnesses such as bacteremia, catheter-related infections, surgical site infections, ventilator-associated pneumonia, otitis, and urinary tract infections [1]. P. aeruginosa is naturally resistant to many common antibiotics due to its reduced expression of high-permeability porins and the acquisition of new resistance determinants. As a result of multi-drug resistance (MDR), infections due to these bacteria became challenging to treat [2], leading to infection outbreaks, longer hospital stays, and higher mortality rates [3].
Antimicrobial resistance (AMR) is one of the major health threats worldwide. It is estimated that bacterial AMR was directly responsible for 1.27 million global deaths in 2019 [4]. The development of MDR limits the treatment options since bacteria could display resistance to a broad spectrum of antibiotic classes, including aminoglycosides (amikacin, gentamicin, and tobramycin), fluoroquinolones (ciprofloxacin, ofloxacin, and norfloxacin), carbapenems, tetracyclines [5], and colistin [6]. Carbapenem antibiotics are the last resort for therapy for infections caused by MDR-P. aeruginosa; however, increased carbapenem-resistant P. aeruginosa (CRPA) has been reported [7,8]. Those mechanisms of carbapenem resistance include efflux of the drug mediated by overexpression of the MexAB-OprM efflux pump, overproduction of AmpC beta-lactamase, inactivation of the OprD outer membrane protein, and production of carbapenemases [9]. Class B carbapenemases or metallo-β-lactamases (MBLs) are major determinants of transferable resistance and include Verona integron-encoded MBLs (VIM) and imipenemase enzymes (IMP) [10]. In Mexico, blaVIM, blaIMP, and blaGES are the most frequent carbapenemase-encoding genes found in P. aeruginosa [11,12,13].
Fluoroquinolones are widely used to treat infections caused by this bacterium. Quinolone resistance is primarily caused by a chromosomal mutation in the quinolone resistance-determining region (QRDR) of the DNA gyrase (gyrA and gyrB) and topoisomerase (parC and parE) encoding genes [14]. However, plasmid-mediated quinolone resistance genes (PMQR) have also been detected in P. aeruginosa [15,16,17,18,19,20,21,22,23,24]. PMQR-mediated mechanisms include the following: qnr genes, that encode the Qnr protein family (QnrA, QnrB, QnrS, QnrC, QnrD, QnrE, and QnrVC) [14]; the acetyltransferase aac-(6′)-lb-cr, which is a variant of an enzyme involved in aminoglycoside acetylation; and the active efflux pumps such as QepA and OqxA [14,21]. PMQR determinants can spread vertically and horizontally among bacteria and facilitate the selection of resistant mutants conferring only low-level fluoroquinolone resistance, which may not be detected by standard susceptibility testing but may increase the frequency of mutations in the chromosomal genes, facilitating the selection of high-level resistance to this antibiotic class [25].
Indeed, quinolone resistance determinant QnrVC has also been linked to some epidemic strains with acquired carbapenemases, such as ST175 and ST244 [26]. Furthermore, the presence of these genes might affect the minimal inhibitory concentration (MIC) to unrelated agents such as novobiocin, tigecycline, or colistin [27]. In addition, several authors have suggested that the presence of the qnr genes is associated with a trend related to extended hospital stays and increased 30-day mortality [27,28]. Thus, the co-occurrence of encoding carbapenemase-genes and PMQR determinants may facilitate the emergence of MDR isolates [29], amplifying antibiotic resistance and limiting the effectiveness of empirical therapy, resulting in the need for more potent and potentially toxic antibiotics such as polymyxins [30]. Additionally, novel agents like ceftazidime-avibactam, ceftolozane-tazobactam, imipenem-cilastatin-relebactam, and cefiderocol may be required [31], however, more clinical trials involving the news therapies are needed to answer these questions.
Given the limited data on the co-existence of PMQR and carbapenemase-encoding genes among clinical isolates of P. aeruginosa worldwide [22,23,24], and particularly in Mexico [32], the characterization of the carbapenemase-encoding genes and evaluation of PMQR determinants in P. aeruginosa isolates from nosocomial patients in Aguascalientes, Mexico, was carried out. Moreover, the colistin resistance mcr-1 gene was also assessed.
2. Materials and Methods
2.1. Sampling and Bacterial Isolation
A total of 52 bacteria isolates were collected from July 2019 to February 2023 from nosocomial patients at Centenario Hospital Miguel Hidalgo, Aguascalientes, Mexico. This was a retrospective and observational study from a single center. Culture specimens were categorized according to their source such as blood, urine, respiratory, or biopsy. In case of a relapse, only the first bacterial isolation was considered. All the isolates were positive for P. aeruginosa by biochemical testing. As a confirmatory test, all the strains were confirmed by PCR as Pseudomonas sp., using the primers of PG-GS-F and PG-GS-R that mark the location 95–113 and 693–712 of the 16SrDNA sequence in Pseudomonas sp., as previously described [33]. P. aeruginosa ATCC 27853 was used as positive control and water as negative control. Primers used are listed in Supplementary Table S1.
2.2. Ethics Statement
The Centenario Hospital Miguel Hidalgo’s Ethics Committee approved this study on 16 January 2023, with the assigned number CEI-CI/008/23.
2.3. Data Collection
Clinical data were obtained from Centenario Hospital Miguel Hidalgo’s electronic clinical record. The repository was used to obtain clinical settings at the time of the culture, microbiology data, comorbidities, length of hospital stay from the day of the index culture (LOS), and complications of patients such as septic shock, mechanical ventilation, and previous antibiotic exposure (any empiric antibiotic prescribed 90 days before the positive culture).
2.4. Antibiotic Susceptibility Testing
All the strains were tested for antimicrobial susceptibility using the Vitek-2 analysis system (bioMérieux, Salt Lake City, UT, USA), according to the manufacturer’s instructions. The antibiotics tested were amikacin, cefepime, ceftazidime, ceftriaxone, ciprofloxacin, gentamicin, imipenem, meropenem, doripenem, piperacillin/tazobactam, colistin, and tigecycline. The MIC breakpoint values for each antibiotic are shown in Supplementary Table S2, CLSI 2020 [34]. However, not all the strains were tested against all the antimicrobial agents since not all the antibiotics were available at the hospital at the time of isolation. Bacteria strains were classified as carbapenem-resistant P. aeruginosa (N = 52) when the bacteria were non-susceptible to imipenem, meropenem, or doripenem [34]. Strains displaying minimum inhibitory concentration (MIC) values of ≥8 µg/mL for carbapenems were considered resistant. Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as quality-control strains. The results were interpreted as susceptible (S), intermediate (I), or resistant (R). The isolates were defined as MDR, extensively drug-resistant (XDR), or pan drug-resistant (PDR) using the clinical laboratory antimicrobial susceptibility testing results and according to a consensus definition [35]. Only carbapenem-resistant P. aeruginosa isolates were included in the study.
2.5. Screening and Identification of PMQR and Carbapenemase-Encoding Genes
For DNA extraction, bacteria were cultured on blood agar for 24 h at 37 °C. The culture was transferred into a sterile Broth Heath Infusion, BHI (BD, Le Pont de Claix, France) 5 mL tube, followed by incubation at 37 °C overnight. One point five mL of the bacterial culture were transferred to microcentrifuge tubes and the cells were pelleted at 12,000 rpm for 5 min. The supernatant was discarded, and the pellet was used for DNA isolation as described by Sambrook and Russell 2001 [36]. The DNA was stored at −20 °C until its use.
First, all the strains were analyzed for the genes encoding the following: (1) serino-β-lactamases (blaGES and blaKPC); (2) metallo-β-lactamases (blaIMP, blaVIM, and blaNDM), and (3) oxacillinases (blaOXA-23, blaOXA-48, blaOXA-51, and blaOXA-1) [37,38]. The amplification programs were performed at 94 °C for 5 min, followed by 35 cycles. Each cycle consisted of 94 °C for 30 s, with various annealing conditions (Supplementary Table S1) for 45 s, and 72 °C for 1 min, with a final extension step of 72 °C for 10 min. Then, a screening for PMQR genes was performed as previously described [39,40,41,42,43]. The screening includes the detection of qnrA, qnrB, qnrS, qnrC, and qnrD, as well as the oqxA and acc-(6′)-lb genes. J53pMG252 strain was used as qnrA positive control, J53pMG298 as qnrB control, J53pMG306 as qnrS control, and Salmonella SA20042859 as a positive control to acc-(6′)-lb. In some cases, previous clinical strains carrying the different target genes were used as positive controls, and water as a negative control. The colistin resistance (mcr-1) gene was also tested. The amplification programs were performed at 94 °C for 5 min, followed by 35 cycles. Each cycle consisted of 94 °C for 30 s, 55°C for 45 s, and 72 °C for 30 s, with a final extension step of 72 °C for 10 min. Primers used are listed in Supplementary Table S1. The amplification products were visualized in agarose gel at 1.5%.
2.6. Statistical Analysis
Statistical data analysis was performed using GraphPad Prim Software (Boston, MA, USA, version 10.0.3). The results were described as descriptive statistics in terms of relative frequency. Comparisons among groups were made using the chi-square, Fisher’s exact test for the categorical data, or the Mann–Whitney U test for the continuous data. Univariate analysis of risk factors for 30-day mortality after positive carbapenem-resistant P. aeruginosa culture was carried out. p-values less than 0.05 were considered statistically significant.
3. Results
3.1. Identification of the Isolates and Clinical Characteristics
Based on a standard bacteriological test, 52 clinical strains were collected overall. All the isolates were positive for P. aeruginosa by biochemical and PCR testing. The clinical characteristics of the patients and the results of the univariate analysis of risk factors for 30-day mortality are shown in Table 1. Initial diagnosis or service of the patients include the following: viral pneumonia (20 isolates, 38.5%), Coronavirus (COVID) (6 isolates, 11.5%), acute respiratory failure (3 isolates, 5.8%), septic shock (2 isolates, 3.8%), kidney stone (3 isolates, 5.8%), woman surgery (2 isolates, 3.8%), septic arthritis (1 isolate, 1.9%), chronic obstructive pulmonary disease (1 isolate, 1.9%), multiple burns (1 isolate, 1.9%), general therapy (5 isolates, 9.6%), encephalopathy (1 isolate, 1.9%), and other conditions (7 isolates, 13.5%). The cases included 28 (53.8%) males and 24 (46.2%) females. The isolates were distributed among all the age groups, with 19 (36.5%) isolates belonging to the age category between 20 and 49 years old, 16 (30.8%) isolates between 50 and 64 years old, and 17 (32.7%) isolates ≥ 65 years old. Most isolates were collected from the respiratory tract (bronchial discharge culture, 34 isolates, 63.5%), followed by urine culture (13 isolates, 25%). Septic shock (OR = 0.19; 95% CI 0.04–0.79, p = 0.023) and infection with XDR bacteria (OR = 0.17; 95% CI 0.03–0.88, p = 0.035) were independent risk factors for 30-day mortality after the culture. Furthermore, 12 strains (23.1%) were isolated from patients with co-infections, including co-infection with Acinetobacter baumannii (4 isolates, 7.7%), Klebsiella pneumoniae (3 isolates, 5.8%), Staphylococcus aureus (1 isolate, 1.9%), Chyseobacterium (F.) indologenes (2 isolates, 3.8%), Candida albicans (1 isolate, 1.9%), and Candida tropicals (1 isolate, 1.9%).

Table 1.
Univariate analysis of risk factors for the 30-day mortality in patients with carbapenem-resistant P. aeruginosa.
3.2. Antimicrobial Susceptibility
Colistin was the most active agent tested for P. aeruginosa infection, with 81.8% susceptibility (Table 2). All the strains exhibited resistance to imipenem (MIC values ranging from 8 to ≥16 µg/mL), meropenem (MIC values ranging from 8 to ≥16 µg/mL), doripenem (MIC values ≥ 8 µg/mL), ceftriaxone (MIC values ranging from 32 to ≥64 µg/mL), and tigecycline (MIC values ≥ 8 µg/mL). In addition, 15 of 52 isolates (28.8%) were categorized as MDR, 30 isolates (57.7%) as XDR, and seven isolates (13.5%) as PDR. Interestingly, all XDR strains were colistin-susceptible. Individual results are described in Supplementary Table S3.

Table 2.
Antimicrobial susceptibility of P. aeruginosa isolates (N = 52).
To determine if there were some differences in the antibiotic-resistant profiles of the isolates based on the susceptibility to quinolones, it was compared to ciprofloxacin-resistant isolates vs. ciprofloxacin-susceptible isolates (Table 3). Antibiotic resistance rates between the two groups were significantly different. Notably, ciprofloxacin-susceptible isolates had higher rates of susceptibility to several antibiotics, including cefepime (50%, p = 0.0018), ceftazidime (33.3%, p = 0.0046), amikacin (100%, p < 0.0001), and gentamicin (100%, p < 0.0001). Based on these findings, it is suggested that ciprofloxacin may be a viable treatment option for this particular phenotype.

Table 3.
Antibiotic resistance rates of ciprofloxacin-resistant and ciprofloxacin-susceptible P. aeruginosa.
3.3. Occurrence of Carbapenemase Encoding-Genes
Out of 52 isolates, 35 (67.3%) were positive for at least one carbapenemase-encoding gene tested. Two isolates (3.8%) were positive for blaIMP; three (5.8%) were positive for blaKPC and blaNDM; four (7.7%) were positive for blaOXA-48, and eight (15.4%) were positive for blaOXA-1 and blaVIM. blaGES was detected only in one isolate (1.9%), and blaOXA-51 was the most frequently carbapenemase-encoding gene found within 22 isolates (42.3%). None of the isolates tested carried blaOXA-23. Seventeen isolates (32.7%) were negative for carbapenemase-encoding genes, suggesting that other mechanisms of resistance to carbapenems are important for the strains isolated at the hospital. Moreover, 38.5% (20 isolates) were positive for only one carbapenemase-encoding gene, 26.9% (14 isolates) co-carrying two different genes, and 1.9% (1 isolate) co-carrying three different genes (blaOXA-51, blaOXA-1, and blaVIM). The distribution of carbapenemase-encoding genes found among isolates is shown in Table 4.

Table 4.
Distribution of carbapenem-encoding genes and PMQR determinants in Pseudomonas aeruginosa isolates (N = 52).
3.4. Screening for Plasmid-Mediated Colistin-Resistant mcr-1 Gene
Only one isolate (1.9%) was positive for mcr-1 (PHH12 isolate, Supplementary Table S4). This strain also carried blaOXA-48 and qnrS genes, was colistin intermediate with MIC values ≤ 0.5 µg/mL, ciprofloxacin-resistant (MIC ≥ 4 µg/mL), doripenem resistant (MIC ≥ 8 µg/mL), imipenem resistant (MIC ≥ 16 µg/mL), and meropenem resistant (MIC ≥ 16 µg/mL). Furthermore, this strain was susceptible to amikacin (MIC = 8). Interestingly, none of the three isolates resistant to colistin (MICs values of ≥16 µg/m and 4 µg/mL) were positive for the mcr-1 gene, indicating the presence of other mechanisms of resistance such as chromosome-encoded mutations.
3.5. Presence of PMQR Determinants
Among the 52 isolates, 18 (34.6%) isolates were positive for the qnr genes, 24 (46.2%) isolates were positive for the oqxA gene, and 13 (25%) for the aac-(6′)-lb gene (Table 4). The results indicated that oqxA was the most prevalent quinolone-resistance gene, followed by the aac-(6′)-lb gene. In the case of qnr genes, qnrS was the most frequent gene (9 isolates, 17.3%), followed by qnrC (7 isolates, 13.5%) and qnrB (2 isolates, 3.8%). qnrD and qnrA were not found in the isolates (Supplementary Table S4).
3.6. Co-Occurrence of Carbapenemase-Encoding Genes and PMQR Determinants
The co-existence of carbapenemase-encoding and quinolone-resistant genes was found among CIP-resistant and CIP-sensible isolates (Table 4). The patterns of co-existence were recognized as follows: blaOXA-51 co-carrying oqxA (11 isolates, 21.1%); blaOXA-51 co-carrying aac-(6´)-lb (6 isolates, 11.5%); blaVIM co-carrying aac-(6′)-lb (2 isolates, 3.8%); blaKPC co-carrying oqxA (3 isolates, 5.8%), with two of them also carrying qnrS; and blaOXA-48 co-carrying qnrS (3 isolates, 5.8%), with two of them also carrying oqxA. However, it seems that the co-existence is unrelated to a specific pattern of ciprofloxacin-resistant determinants. No significant differences were found between PMQR and ciprofloxacin-resistant and sensitive isolates.
4. Discussion
The incidence of plasmid-mediated quinolone-resistant determinants and carbapenemase-encoding genes simultaneously in P. aeruginosa has been analyzed in limited studies. In this study, the prevalence of PMQR and carbapenemase-encoding genes occurring at the same time in P. aeruginosa strains isolated from clinical patients in Aguascalientes, Mexico was reported. Fifty-two strains were investigated. The highest proportion of carbapenem-resistant P. aeruginosa was isolated from respiratory tract samples. This was in concordant with previous studies since carbapenem resistance is directly influenced by the culture site, being higher in respiratory infections than in bloodstream infections [44]. Additionally, urine cultures had a great proportion of carbapenem-resistant P. aeruginosa isolates in this study. Moreover, we found that septic shock was an independent risk factor for 30-day mortality as well as extensively drug-resistant P. aeruginosa (XDR-PA), in agreement with previous studies [45,46,47].
Higher resistance levels to all tested antibiotics were identified in the study by Nieto-Saucedo et al. [13]. In the comparative analysis, resistance rates were as follows: amikacin (24% vs. 73.1%), gentamicin (60% vs. 75%), ceftazidime (52% vs. 83.3%), cefepime (44% vs. 76%), ciprofloxacin (44% vs. 88.4%), colistin (0% vs. 6.8%), and piperacillin/tazobactam (36% vs. 71.8%). This could be due to geographic variation, healthcare practices, and the genetic mechanisms underlying resistance. Conversely, the study by Martinez-Zavaleta et al. [48], also from Mexico, reported similar resistance rates in comparison to our findings, showing resistance levels for amikacin (50% vs. 73.1%), gentamicin (70.8% vs. 75%), ceftazidime (73.9% vs. 83.3%), cefepime (76.5% vs. 76%), colistin (9.9% vs. 6.8%), and piperacillin/tazobactam (57.3% vs. 71.8%). Furthermore, a study in Nigeria [49] reported resistance rates of approximately 75% for ceftazidime, cefepime, ciprofloxacin, and piperacillin/tazobactam in CRPA. Another study in Japan found that 47.1% of the CRPA isolates were resistant to piperacillin-tazobactam and 73.4% to ciprofloxacin [50], which aligns with the antimicrobial susceptibility found in this study.
Additionally, colistin (polymyxin E) was the most active agent tested, with less than seven percent of the isolates resistant, suggesting its use as a better alternative to treat these infections. The prevalence of colistin resistance was higher than one reported in a previous study in Mexico (1.65% of prevalence) [51]. Even though colistin seems to be the best option for treatment due to a lower resistance rate, this antibiotic could cause harmful effects on human health, and precautions must be taken regarding its use. There were notable variations in antibiotic resistance rates between the ciprofloxacin-resistant and ciprofloxacin-susceptible isolates, which differed from a previous study [15]. Furthermore, the ciprofloxacin-susceptible isolates exhibited higher susceptibility to aminoglycosides, ceftazidime, and cefepime, suggesting a viable alternative for this type of phenotype. Similarly, in a previous study [45], all XDR-P. aeruginosa isolates were susceptible to colistin, suggesting its use as a better alternative treatment.
This study included only the CRPA isolates and the first eligible culture episode for each patient. This approach was taken to minimize the chances of including the same isolate multiple times, as some patients experienced two or three reinfections during the study period. Moreover, the period of the study (2019 to 2023), the diversity of the specimen types (blood, biopsy, respiratory tract, and urine), and the genes found in the different isolates suggest that they represent distinct bacterial strains.
Among carbapenemase-encoding genes, Nieto-Saucedo et al. [13] found the blaIMP-75 gene as the most common carbapenemase-encoding gene for P. aeruginosa, contrasting with our study since we detected only two isolates carrying blaIMP genes. The genes blaGES and blaVIM were also found in P. aeruginosa [13,32]. In our study, blaGES and blaVIM were also detected in the isolates, with blaVIM and blaOXA-51 being the most frequently detected carbapenemase-encoding genes, which agrees with previous studies in Mexico, where blaVIM resulted in the primary gene reported in the same bacteria [11]. Detection of blaOXA-51 is rare for P. aeruginosa since this gene encoding Ambler class D oxacillinase (OXA)-like carbapenemase, blaOXA-51 is naturally detected in A. baumannii and is related in resistance to oxacillin, cephalosporins, and carbapenems [52,53]. However, in the last years, it has been emerging in P. aeruginosa [54], as well as other oxacillinase-type enzymes such as blaOXA-48 [55,56]. Moreover, we included various types of hospital-acquired infections caused by P. aeruginosa such as monomicrobial and polymicrobial infections. Indeed, several strains were isolated from patients with co-infection by A. baumannii, thus facilitating the spread of resistant determinants through horizontal gene transfer, as well as influencing the treatment responses and patient outcomes.
Carbapenemase-encoding genes were not amplified in thirty-two percent of the isolates. Therefore, the production of carbapenemases may not be the primary mechanism of resistance in P. aeruginosa strains circulating in the hospital, which is closer to those described by Garza-Ramos et al. [11], where 47% of the isolates were negative for the presence of the carbapenemase-encoding genes tested. Other mechanisms may include alteration or lack of porin OprD which has been related to reduced susceptibility to carbapenems [57], overexpression of efflux pumps, mutations in AmpC and its regulatory genes, and plasmid-encoded AmpC. Nevertheless, detecting MBL-producing strains is of great importance since options for P. aeruginosa are still limited. Lately, the treatment suggested for MBL-carbapenemase includes cefiderocol, fosfomycin, high-dose amikacin, and a synergic combination of colistin with fosfomycin or aminoglycoside, or the combination of ceftolozane-tazobactam with amikacin or fosfomycin [58,59]. Meanwhile, ceftazidime-avibactam could be usable for GES-carbapenemase P. aeruginosa [58], KPC and OXA-48 producers; ceftazidime-avibactam plus aztreonam seems to be a reliable option for MLBs producers [31].
Only one strain tested carried the mcr-1 gene. Despite their harmful effects on human health, polymyxins remain a last resort against Gram-negative MDR infections, especially in carbapenem-resistant bacteria. The mcr genes are of great concern due to their high potential for horizontal propagation. Among the variants described, mcr-1 is the most prevalent within Mexico [11] and globally [60]. Previous reports have also revealed the emergence of mcr-1 in P. aeruginosa [61,62]. In Mexico, mcr-1 has also been reported in Klebsiella pneumoneae [11] and Escherichia coli [51]; meanwhile, mcr-2 has been detected in Enterobacter cloacae [13]. The horizontal propagation of the mcr-1 gene into carbapenem-resistant bacteria could severely limit antimicrobial treatment options. Thus, it is essential to strengthen our surveillance of bacteria exhibiting this resistance mechanism.
Among fluoroquinolone resistance genes, there were found frequencies of 46.2% for oqxA, 25% for aac-(6′)-lb, and 34.6% for qnr genes (17.3% for qnrS, 13.5% for qnrC, and 3.8% for qnrB). The qnrD and qnrA genes were not detected. This agrees with Venkataramana et al. [19], who found that plasmid-meditated resistance acc-(6′)-lb-cr is primarily responsible for mediating fluoroquinolone resistance in clinical isolates of P. aeruginosa. However, higher frequency was found within 77.6% of the isolates testing positive for this gene. The authors also found the genes qnrB (14.1%), qnrS (14.1%), and oqxAB (3.5%) in the isolates. Similar results were detected by Abdelrahim et al., who showed a frequency of 77.3% for acc-(6′)-lb-cr gene in P. aeruginosa [63]. In our study, several ciprofloxacin-resistant isolates were negative for the PMQR tested. This might be attributable to the major mechanism for fluoroquinolone resistance in P. aeruginosa, the mutations in the DNA gyrase encoded by the gyrA and gyrB genes, as well as in the topoisomerase IV encoded by the parC and parE genes [64].
Interestingly, in our study, oqxA has the highest frequency (46.2%), which encodes for a multi-drug efflux pump belonging to the resistance-nodulation-division family (RND) [14], implying its contribution to the reduced susceptibility to quinolones in the clinical isolates of the region. In the study by Andres et al. [65], oqxA and oqxB were found in isolates with a wide range of MIC values for ciprofloxacin, including some that were susceptible. Moreover, Goudarzi et al. [66] highlighted that these genes were detected in both susceptible and resistant isolates. This finding is supported by Agyepong et al. [67], who observed that oqxA and oqxB were present together in susceptible and resistant ciprofloxacin isolates. This agrees with our study, where several strains carrying oqxA were ciprofloxacin-sensitive isolates, and the resistant ones have MIC values ≥ 4 µg/mL, suggesting that while oqxA is associated with resistance, it can also coexist with susceptibility or low-level-resistant isolates. Furthermore, the detection of oqxA gene is worrisome since the OqxAB multi-drug efflux pump is linked to low to intermediate resistance to other antibiotics such as quinoxalines, quinolones, tigecycline, nitrofurantoin, chloramphenicol, several detergents and disinfectants [68], and the high level of expression of this efflux pump is associated to high resistant to ciprofloxacin [69]. Additionally, the presence of aac(6′)-lb-cr gene enables the selection of highly ciprofloxacin-resistant chromosomal mutants. It converts the low-level fluoroquinolone resistance mediated by this enzyme into high-level resistance when present alongside Qnr proteins [63].
Like earlier studies [19,70], the qnrA gene was not detected in any of the isolates of P. aeruginosa in this study. In contrast to our study, Lopez-Garcia et al. [32] did not find the genes blaVIM, blaNDM, blaKPC, qnrA, qnrB, qnrS or aac-(6′)-Ib-cr in MDR P. aeruginosa strains. Regardless, blaIMP (41%), blaGES (49%), blaOXA-2 (85%), and blaOXA-50 (100%) were found. Another study in Iran [15] found higher prevalence rates of qnr genes compared to our study: qnrA (25.8% vs. 0%), qnrB (29.2% vs. 3.8%), and qnrS (20.8% vs. 17.3%); however, qnrD was also not detected as in this study.
Although some studies were carried out to detect PMQR in P. aeruginosa, recent reports suggest a current increase in the mobile genetic pool in P. aeruginosa which could provide multiple resistance to the most clinically used antibiotics [71]. Indeed, qnrS1 was detected alongside the β-lactamase gene blaTEM-1 [72], while qnrVC was detected with the blaVIM-2 gene as part of class 1 integron [73]. Elena et al. [22] found the simultaneous presence of blaVIM-11 and the PMQR qnrS1 in the same strain located in different plasmids. Other studies have shown evidence of qnrVC1 and blaNDM-1 in the high-risk P. aeruginosa ST773 clone reported worldwide as MDR [24]. Lopez-Garcia et al. [32] found blaIMP-18 + aacA7 + blaIMP-62 + qacH + aacA4 + aadA1 + blaOXA-2 in two strain isolates from the same patient. Similarly, the co-existence of resistance genes in different combinations in P. aeruginosa has also been previously reported with multiple ESBL genes and carbapenemase-encoding genes (CTX-M-1, NDM-1, and OXA-48), with the addition of acc-(6′)-lb [74].
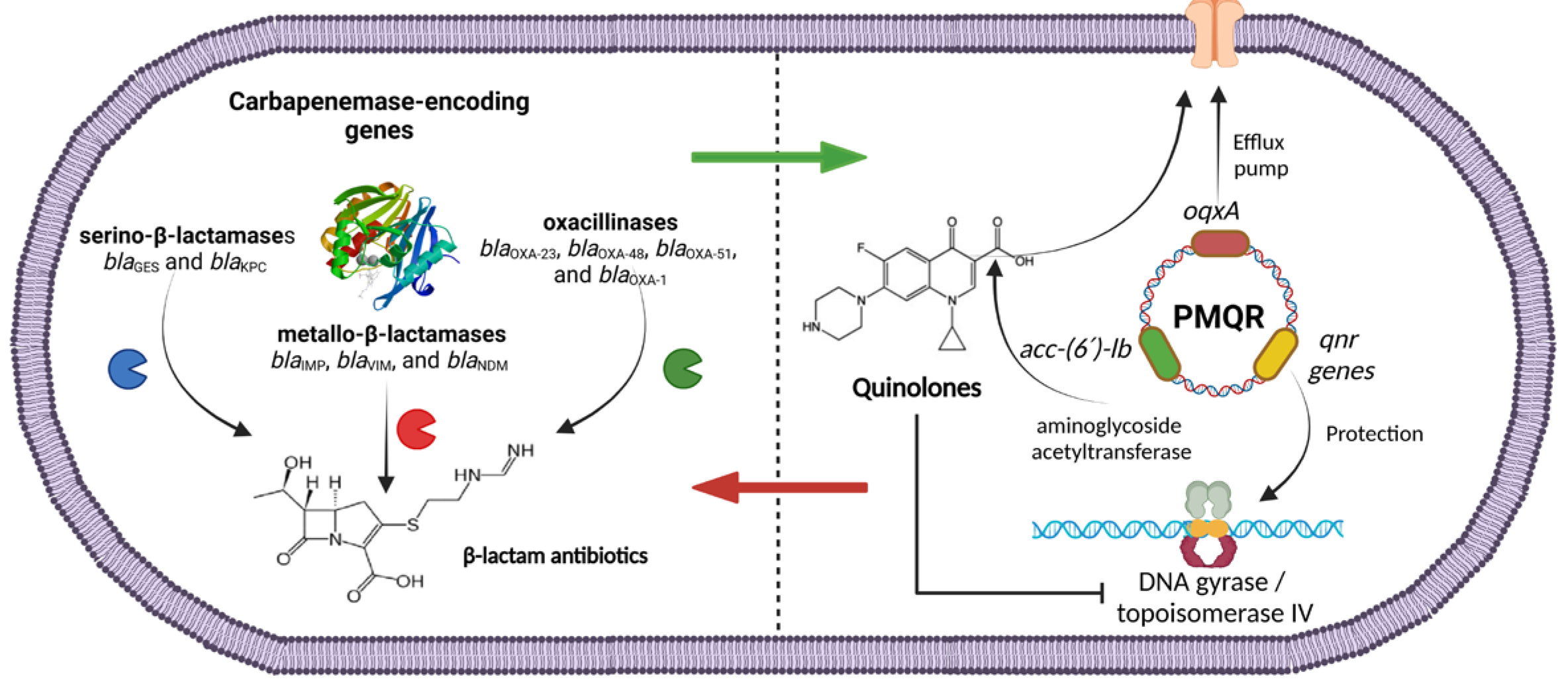
In this study, it was found that PMQR was present on resistant and susceptible ciprofloxacin isolates, indicating a possible future increase in quinolone resistance. Several isolates carried carbapenemase-encoding genes, and PMQR determinants simultaneously were found, indicating the dissemination of PMQR on carbapenem-resistant strains. Figure 1 describes the presence of both genetic elements in P. aeruginosa. OXA-type carbapenemase-encoding genes were found in combination with efflux pumps and aminoglycoside variants (blaOXA-51 + oqxA + aac-(6′)-lb) as well as serin-based carbapenemase-encoding genes with qnr-encoding variants and efflux pump genes (blaKPC + qnrB, qnrC, qnrS + oqxA; blaGES + qnrB, qnrS + oqxA). The co-carriage of carbapenemase-encoding genes and PMQR determinants might suggest the presence of circulating plasmids that carry these resistance genes [63]. P. aeruginosa is a highly diverse pathogen that can adapt to its environment. The rapid development of antimicrobial resistance in this bacterium may be attributed to the excessive and inappropriate use of antibiotics. This creates selective pressure which can lead to mutations, the acquisition of resistance genes through horizontal gene transfer, the overexpression of beta-lactamases, and the proliferation of antibiotic resistance [75]. While PMQR determinants by themselves generally produce low levels of fluoroquinolone resistance, this plays a significant role in the emergence of clinical resistance to ciprofloxacin. This allows the bacteria to grow at clinically relevant concentrations of quinolones, potentially leading to treatment failures.
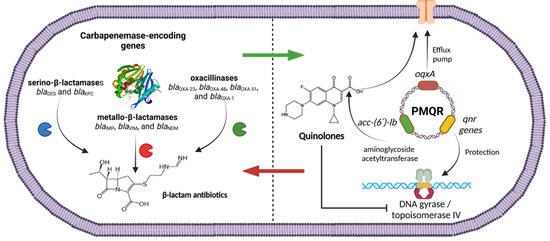
Figure 1.
Plasmid-mediated quinolone-resistance genes (PMQR) and carbapenemase-encoding genes found in CRPA isolates in this study.
The frequency of PMQR genes in clinical isolates of carbapenem-resistant P. aeruginosa is worrisome since it enables their spread to other bacterial species by horizontal gene transfer. It is important to emphasize that understanding the mechanisms of carbapenem resistance in clinical isolates of P. aeruginosa is crucial in determining the most effective treatment strategy.
Our study has some limitations. Firstly, all the strains were not tested with all antibiotics because not all antibiotics were available at the time of the isolation. Secondly, the study involved patients in one hospital center. To increase the statistical power of the results, it would be necessary in the future to include a higher number of samples in a larger multicenter study to improve the generalizability of the findings. However, although this study is limited to a single center, it aims to guide the rational prescription of antibiotics in a regional context. Finally, although all isolates come from different patients, the study did not assess the genetic relationships among the resistant strains.
5. Conclusions
To the best of our knowledge, this is the first report in Mexico of PMQR genes in carbapenem-resistant P. aeruginosa. The co-existence of PMQR with carbapenemase-encoding genes in carbapenem-resistant P. aeruginosa is infrequent, though its occurrence could complicate infection treatments and increase resistance through horizontal gene transfer. Routine screenings are advisable as an infection control measure. This study aims to guide the rational prescription of antibiotics in a regional context.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13110992/s1, Table S1: Oligonucleotides used in this study; Table S2: Breakpoint values of the antimicrobial agents used in this study; Table S3: Antimicrobial susceptibility of carbapenem-resistant P. aeruginosa isolates; Table S4: Carbapenemase-encoding and plasmid-mediated quinolone-resistant genes in P. aeruginosa.
Author Contributions
Conceptualization, A.L.G.-B., J.M.A.-G. and M.G.-G.; methodology, A.S.T.-C., F.Y.R.-C., D.E.G.-P., C.L.R.-M., C.A.-Á., F.G.-G., A.C.M.-F. and R.G.-R.; investigation, A.S.T.-C., F.Y.R.-C. and D.E.G.-P.; resources, A.L.G.-B., J.M.A.-G., M.G.-G., F.J.A.-G., A.L.-M. and C.L.R.-M.; data curation, A.S.T.-C., F.Y.R.-C., D.E.G.-P. and A.L.-M.; writing—original draft preparation, A.S.T.-C., A.L.-M. and F.Y.R.-C.; writing—review and editing, A.L.G.-B., F.J.A.-G., E.H.-C. and A.L.-M.; supervision, A.L.G.-B., J.M.A.-G., M.G.-G. and F.J.A.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The internal research project PIB19-3 of the Universidad Autónoma de Aguascalientes supported the study. F.Y.R.C. received a research grant from CONAHCYT (grant number 373176).
Institutional Review Board Statement
This study was approved by the Centenario Hospital Miguel Hidalgo Ethics and Research Committee with the number CEI-CI/008/23 (16 January 2023).
Informed Consent Statement
Patient consent was waived due to the characteristics of cohort research, which is a retrospective and observational study. This study does not involve risk to subjects, and the waiver does not negatively impact their rights or welfare. All the samples were taken for routine analyses to identify nosocomial infections. The hospital considered the approval by the Ethics and Research Committee adequate.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We acknowledge the collaboration and technical support of the Centenario Hospital Miguel Hidalgo Clinical Laboratory at Aguascalientes, Mexico, especially the Bacteriology Section.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Bonomo, R.A.; Szabo, D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56. [Google Scholar] [CrossRef]
- Ramanathan, S.; Fitzpatrick, M.A.; Suda, K.J.; Burns, S.P.; Jones, M.M.; LaVela, S.L.; Evans, C.T. Multidrug-resistant Gram-negative organisms and association with 1-year mortality, readmission, and length of stay in Veterans with spinal cord injuries and disorders. Spinal Cord 2020, 58, 596–608. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Avakh, A.; Grant, G.D.; Cheesman, M.J.; Kalkundri, T.; Hall, S. The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy. Antibiotics 2023, 12, 1304. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huan, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infct. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Wieland, K.; Chhatwal, P.; Vonberg, R.P. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am. J. Infect. Control. 2018, 46, 643–648. [Google Scholar] [CrossRef]
- Huang, W.; Wei, X.; Xu, G.; Zhang, X.; Wang, X. Carbapenem-resistant Pseudomonas aeruginosa infections in critically ill children: Prevalence, risk factors, and impact on outcome in a large tertiary pediatric hospital of China. Front. Public Heal. 2023, 11, 1088262. [Google Scholar] [CrossRef]
- Pai, H.; Kim, J.; Kim, J.; Lee, J.H.; Choe, K.W.; Gotoh, N. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2001, 45, 480–484. [Google Scholar] [CrossRef]
- Cornaglia, G.; Giamarellou, H.; Rossolini, G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011, 11, 381–393. [Google Scholar] [CrossRef]
- Garza-Ramos, U.; Silva-Sánchez, J.; López-Jácome, L.E.; Hernández-Durán, M.; Colín-Castro, C.A.; Sánchez-Pérez, A.; Rodríguez-Santiago, J.; Morfín-Otero, R.; Rodríguez-Noriega, E.; Velázquez-Acosta, M.D.; et al. Carbapenemase-Encoding Genes and Colistin Resistance in Gram-Negative Bacteria During the COVID-19 Pandemic in Mexico: Results from the Invifar Network. Microb. Drug. Resist. 2023, 29, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Garza-Ramos, U.; Morfin-Otero, R.; Sader, H.S.; Jones, R.N.; Hernández, E.; Rodríguez-Noriega, E.; Sánchez, A.; Carrillo, B.; Esparza-Ahumada, S.; Silva-Sánchez, J. Metallo-beta-lactamase gene bla (IMP-15) in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob. Agents Chemother. 2008, 52, 2943–2946. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nieto-Saucedo, J.R.; López-Jacome, L.E.; Franco-Cendejas, R.; Colín-Castro, C.A.; Hernández-Duran, M.; Rivera-Garay, L.R.; Zamarripa-Martinez, K.S.; Mosqueda-Gómez, J.L. Carbapenem-Resistant Gram-Negative Bacilli Characterization in a Tertiary Care Center from El Bajio, Mexico. Antibiotics 2023, 12, 1295. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updates 2016, 29, 13–29. [Google Scholar] [CrossRef]
- Saki, M.; Farajzadeh Sheikh, A.; Seyed-Mohammadi, S.; Asareh Zadegan Dezfuli, A.; Shanin, M.; Tabasi, M.; Veisi, H.; Keshavarzi, R.; Khani, P. Occurrence of plasmid-mediated quinolone resistance genes in Pseudomonas aeruginosa strains isolated from clinical specimens in southwest Iran: A multicentral study. Sci Rep. 2022, 12, 2296. [Google Scholar] [CrossRef]
- Al-Marjani, M.F. Presence of qnr gene in environmental and clinical Pseudomonas aeruginosa isolates in Baghdad. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 853–857. [Google Scholar]
- Yang, X.; Xing, B.; Liang, C.; Ye, Z.; Zhang, Y. Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int. J. Clin. Exp. Med. 2015, 8, 1386–1390. [Google Scholar]
- Liu, J.; Yang, L.; Li, L.; Li, B.; Chen, D.; Xu, Z. Comparative genomic analyses of two novel qnrVC6 carrying multidrug-resistant Pseudomonas. spp strains, Microb. Pathog. 2018, 123, 269–274. [Google Scholar] [CrossRef]
- Venkataramana, G.P.; Lalitha, A.K.V.; Mariappan, S.; Sekar, U. Plasmid-Mediated Fluoroquinolone Resistance in Pseudomonas aeruginosa and Acinetobacter baumannii. J. Lab. Physicians 2022, 14, 271–277. [Google Scholar] [CrossRef]
- Lin, J.; Chen, D.Q.; Hong, J.; Huang, H.; Xu, X. Prevalence of qnrVC Genes in Pseudomonas aeruginosa Clinical Isolates from Guangdong, China. Curr. Microbiol. 2020, 77, 1532–1539. [Google Scholar] [CrossRef]
- Taha, S.A.; Omar, H.H. Characterization of plasmid-mediated qnrA and qnrB genes among Enterobacteriaceae strains: Quinolone resistance and ESBL production in Ismailia, Egypt. Egypt. J. Med. Hum. Genet. 2019, 20, 1–7. [Google Scholar] [CrossRef]
- Elena, A.; Quinteros, M.; Di Conza, J.; Gutkind, G.; Cejas, D.; Radice, M.A. Full characterization of an IncR plasmid harboring qnrS1 recovered from a VIM-11-producing Pseudomonas aeruginosa. Rev. Argent. Microbiol. 2020, 52, 298–304. [Google Scholar] [CrossRef]
- Araujo, B.F.; Ferreira, M.L.; Campos, P.A.; Royer, S.; Batistão, D.W.; Dantas, R.C.; Gonçalves, I.R.; Faria, A.L.; Brito, C.S.; Yokosawa, J.; et al. Clinical and Molecular Epidemiology of Multidrug-Resistant P. aeruginosa Carrying aac (6′)-Ib-cr, qnrS1 and blaSPM Genes in Brazil. PLoS ONE 2016, 11, e0155914. [Google Scholar] [CrossRef]
- Kocsis, B.; Toth, A.; Gulyas, D.; Ligeti, B.; Katona, K.; Rokusz, L.; Szabo, D. Acquired qnrVC1 and blaNDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J. Med. Microbiol. 2019, 68, 336–338. [Google Scholar] [CrossRef]
- Domokos, J.; Kristóf, K.; Szabó, D. Plasmid-mediated quinolone resistance among extended spectrum beta lactase producing Enterobacteriaceae from bloodstream infections. Acta Microbiol. Immunol. Hung. 2016, 63, 313–323. [Google Scholar] [CrossRef]
- Oliver, A.; Rojo-Molinero, E.; Arca-Suarez, J.; Beşli, Y.; Bogaerts, P.; Cantón, R.; Cimen, C.; Croughs, P.D.; Denis, O.; Giske, C.G.; et al. Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2023, 30, 469–480. [Google Scholar] [CrossRef]
- Ruiz, J. Transferable mechanisms of Quinolone Resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, e00007–19. [Google Scholar] [CrossRef]
- Liao, C.H.; Hsueh, P.R.; Jacoby, G.A.; Hooper, D.C. Risk factors and clinical characteristics of patients with qnr-positive Klebsiella pneumoniae bacteraemia. J. Antimicrob. Chemother. 2013, 68, 2907–2914. [Google Scholar] [CrossRef][Green Version]
- Hoseinzadeh, M.; Sedighi, M.; Yahyapour, Y.; Javanian, M.; Beiranvand, M.; Mohammadi, M.; Zarei, S.; Pournajaf, A.; Ebrahimzadeh Namvar, A. Prevalence of plasmid-mediated quinolone resistance genes in extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolates in northern Iran. Heliyon 2024, 10, e37534. [Google Scholar] [CrossRef]
- Yuan, F.; Xiao, W.; Wang, X.; Fu, Y.; Wei, X. Clinical characteristics and prognosis of bloodstream infection with carbapenem-resistant pseudomonas aeruginosa in patients with hematologic malignancies. Infect. Drug Resist. 2023, 16, 4943–4952. [Google Scholar] [CrossRef]
- Aslan, A.T.; Akova, M. Recent updates in treating carbapenem-resistant infections in patients with hematological malignancies. Expert Rev. Anti Infect. Ther. 2024, 1–17. [Google Scholar] [CrossRef] [PubMed]
- López-García, A.; Del Carmen Rocha-Gracia, R.; Bello-López, E.; Juárez-Zelucualtecalt, C.; Sáenz, Y.; Castañeda-Lucio, M.; López-Piego, L.; González-Vázquez, M.C.; Torres, C.; Ayala-Nuñez, T.; et al. Characterization of antimicrobial resistance mechanisms in carbapenem-resistant Pseudomonas aeruginosa carrying IMP variants recovered form a Mexican Hospital. Infect. Drug. Resist. 2018, 11, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Spilker, T.; Coenye, T.; Vandamme, P.; LiPuma, J.J. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004, 42, 2074–2079. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Clinical Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI document M100-ED30; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liligequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning, A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Lariviere, S.; Harel, J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef]
- Cerezales, M.; Biniossek, L.; Gerson, S.; Xanthopoulou, K.; Wille, J.; Wohlfarth, E.; Kaase, M.; Seifert, H.; Higgins, P.G. Novel multiplex PCRs for detection of the most prevalent carbapenemase genes in Gram-negative bacteria within Germany. J. Med. Microbiol. 2021, 70, 3214–3221. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 2006, 50, 2872–2874. [Google Scholar] [CrossRef]
- Wang, M.; Guo, Q.; Xu, X.; Wang, X.; Ye, X.; Wu, S.; Hooper, D.C.; Wang, M. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolates of Proteus mirabilis. Antimicrob. Agents Chemother. 2009, 53, 1892–1897. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Hasman, H.; Xia, S.; Aarestrup, F.M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 2009, 53, 603–608. [Google Scholar] [CrossRef]
- Park, C.H.; Robicsek, A.; Jacoby, G.A.; Sahm, D.; Hooper, D.C. Prevalence in the United States of aac (6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006, 50, 3953–3955. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, W.; Pan, W.; Yin, J.; Pan, Z.; Gao, S.; Jiao, X. Prevalence of qnr, aac (6′)-Ib-cr, qepA, and oqxAB in Escherichia coli Isolates from Humans, Animals, and the Environment. Antimicrob. Agents Chemother. 2012, 56, 3423–3427. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Echols, R.; Magee, G.; Arjona Ferreira, J.C.; Morgan, G.; Ariyasu, M.; Sawada, T.; Nagata, T.D. Prevalence of Carbapenem-Resistant Gram-Negative Infections in the United States Predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect. Dis. 2017, 4, ofx176. [Google Scholar] [CrossRef] [PubMed]
- Palavutitotai, N.; Jitmuang, A.; Tongsai, S.; Kiratisin, P.; Angkasekwinai, N. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS ONE 2018, 13, e0193431. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jun, Y.H.; Kim, Y.R.; Park, K.G.; Park, Y.J.; Kang, J.Y.; Kim, S.I. Risk factors for mortality in patients with Pseudomonas aeruginosa bacteremia; retrospective study of impact of combination antimicrobial therapy. BMC Infect. Dis. 2014, 14, 161. [Google Scholar] [CrossRef] [PubMed]
- Frem, J.A.; Doumat, G.; Kazma, J.; Gharamti, A.; Kanj, S.S.; Fayad, A.G.A.; Matar, G.M.; Kanafani, Z.A. Clinical predictors of mortality in patients with Pseudomonas aeruginosa infection. PLoS ONE 2013, 18, e0282276. [Google Scholar] [CrossRef]
- Martínez-Zavaleta, M.G.; Fernández-Rodríguez, D.; Hernández-Durán, M.; Colín-Castro, C.A.; García-Hernández, M.d.L.; Becerra-Lobato, N.; Franco-Cendejas, R.; López-Jácome, L.E. Acquired blaVIM and blaGES Carbapenemase-Encoding Genes in Pseudomonas aeruginosa: A Seven-Year Survey Highlighting an Increasing Epidemiological Threat. Pathogens 2023, 12, 1256. [Google Scholar] [CrossRef]
- Kawa, D.E.; Tickler, I.A.; Tenover, F.C.; Shettima, S.A. Characterization of Beta-Lactamase and Fluoroquinolone Resistance Determinants in Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolates from a Tertiary Hospital in Yola, Nigeria. Trop. Med. Infect. Dis. 2023, 8, 500. [Google Scholar] [CrossRef]
- Yano, H.; Hayashi, W.; Kawakami, S.; Aoki, S.; Anzai, E.; Zuo, H.; Kitamura, N.; Hirabayashi, A.; Kajihara, T.; Kayama, S.; et al. Nationwide genome surveillance of carbapenem-resistant Pseudomonas aeruginosa in Japan. Antimicrob. Agents Chemother. 2024, 68, e0166923. [Google Scholar] [CrossRef]
- Galindo-Méndez, M.; Navarrete-Salazar, H.; Pacheco-Vásquez, R.; Quintas-de la Paz, D.; Baltazar-Jiménez, I.; Santiago-Luna, J.D.; Guadarrama-Monroy, L. Detection of Plasmid-Mediated Resistance against Colistin in Multi-Drug-Resistant Gram-Negative Bacilli Isolated from a Tertiary Hospital. Microorganisms 2023, 11, 1996. [Google Scholar] [CrossRef]
- Xiao, S.Z.; Chu, H.Q.; Han, L.Z.; Zhang, Z.M.; Li, B.; Zhao, L.; Xu, L. Resistant mechanisms and molecular epidemiology of imipenem-resistant Acinetobacter baumannii. Mol. Med. Rep. 2016, 14, 2483–2488. [Google Scholar] [CrossRef][Green Version]
- Uwingabiye, J.; Lemnouer, A.; Roca, I.; Alouane, T.; Frikh, M.; Belefquih, B.; Bssaibis, F.; Maleb, A.; Benlahlou, Y.; Kassouati, J.; et al. Clonal diversity and detection of carbapenem resistance encoding genes among multidrug-resistant Acinetobacter baumannii isolates recovered from patients and environment in two intensive care units in a Moroccan hospital. Antimicrob. Resist. Infect. Control. 2017, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Nitz, F.; de Melo, B.O.; da Silva, L.C.N.; de Souza Monteiro, A.; Marques, S.G.; Monteiro-Neto, V.; de Jesus Gomes Turri, R.; Junior, A.D.S.; Conceição, P.C.R.; Magalhães, H.J.C.; et al. Molecular Detection of Drug-Resistance Genes of blaOXA-23-blaOXA-51 and mcr-1 in Clinical Isolates of Pseudomonas aeruginosa. Microorganisms 2021, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Gondal, A.J.; Choudhry, N.; Niaz, A.; Yasmin, N. Molecular Analysis of Carbapenem and Aminoglycoside Resistance Genes in Carbapenem-Resistant Pseudomonas aeruginosa Clinical Strains: A Challenge for Tertiary Care Hospitals. Antibiotics 2024, 13, 191. [Google Scholar] [CrossRef] [PubMed]
- Tarafdar, F.; Jafari, B.; Azimi, T. Evaluating the antimicrobial resistance patterns and molecular frequency of blaOXA-48 and blaGES-2 genes in Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from burn wound infection in Tehran, Iran. New Microbes New Infect. 2020, 37, 100686. [Google Scholar] [CrossRef] [PubMed]
- Wolter, D.J.; Hanson, N.D.; Lister, P.D. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol. Lett. 2004, 236, 137–143. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Galani, I.; Papoutsaki, V.; Karantani, I.; Karaiskos, I.; Galani, L.; Adamou, P.; Deliolais, I.; Kodonaki, A.; Papadogeogarki, E.; Markopoulou, M.; et al. In vitro activity of ceftolozane/tazobactam alone and in combination with amikacin against MDR/XDR Pseudomonas aeruginosa isolates from Greece. J. Antimicrob. Chemother. 2020, 75, 2164–2172. [Google Scholar] [CrossRef]
- Nang, S.C.; Li, J.; Velkov, T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef]
- El-Baky, R.M.A.; Masoud, S.M.; Mohamed, D.S.; Waly, N.G.; Shafik, E.; A Mohareb, D.; Elkady, A.; Elbadr, M.M.; Hetta, H.F. Prevalence and Some Possible Mechanisms of Colistin Resistance among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2020, 13, 323–332. [Google Scholar] [CrossRef]
- Snesrud, E.; Maybank, R.; Kwak, Y.I.; Jones, A.R.; Hinkle, M.K.; McGann, P. Chromosomally Encoded mcr-5 in Colistin-Nonsusceptible Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e00679-e18. [Google Scholar] [CrossRef]
- Abdelrahim, S.S.; Hassuna, N.A.; Waly, N.G.F.M.; Kotb, D.N.; Abdelhamid, H.; Zaki, S. Coexistence of plasmid-mediated quinolone resistance (PMQR) and extended-spectrum beta-lactamase (ESBL) genes among clinical Pseudomonas aeruginosa isolates in Egypt. BMC Microbiol. 2024, 24, 175. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Ahangarzadeh Rezaee, M.; Hasani, A.; Aghazadeh, M.; Asgharzadeh, M. The role of gyrA and parC mutations in fluoroquinolones-resistant Pseudomonas aeruginosa isolates from Iran. Braz. J. Microbiol. 2016, 47, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Andres, P.; Lucero, C.; Soler-Bistué, A.; Guerriero, L.; Albornoz, E.; Tran, T.; Zorreguieta, A.; PMQR Group; Galas, M.; Corso, A.; et al. Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob. Agents Chemother. 2013, 57, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, M.; Azad, M.; Seyedjavadi, S.S. Prevalence of plasmid-mediated quinolone resistance determinants and oqxab efflux pumps among extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from patients with nosocomial urinary tract infection in Tehran, Iran. Scientifica 2015, 2015, 1–7. [Google Scholar] [CrossRef]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Amoako, D.G.; Allam, M.; Janice, J.; Pedersen, T.; Sundsfjord, A.; Essack, S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019, 85, 117–123. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Ning, J.; Sajid, A.; Cheng, G.; Yuan, Z.; Hao, H. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control. 2019, 8, 44. [Google Scholar] [CrossRef]
- Amereh, F.; Arabestani, M.R.; Shokoohizadeh, L. Relationship of OqxAB efflux pump to antibiotic resistance, mainly fluoroquinolones in Klebsiella pneumoniae, isolated from hospitalized patients. Iran. J. Basic Med. Sci. 2023, 26, 93–98. [Google Scholar] [CrossRef]
- Nazik, H.; Ongen, B.; Kuvat, N. Investigation of plasmid-mediated quinolone resistance among isolates obtained in a Turkish intensive care unit. Jpn. J. Infect. Dis. 2008, 61, 310–312. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005, 56, 463–469. [Google Scholar] [CrossRef]
- Jiang, X.; Yu, T.; Jiang, X.; Zhang, W.; Zhang, L.; Ma, J. Emergence of plasmid-mediated quinolone resistance genes in clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa in Henan, China. Diagn. Micr. Infec. Dis. 2014, 79, 381–383. [Google Scholar] [CrossRef]
- Belotti, P.T.; Thabet, L.; Laffargue, A.; André, C.; Coulange-Mayonnove, L.; Arpin, C.; Messadi, A.; M’ Zali, F.; Quentin, C.; Dubois, V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015, 70, 2237–2240. [Google Scholar] [CrossRef] [PubMed]
- Sarjana Safain, K.; Bhuyan, G.S.; Hassan Hasib, S.; Islam, M.S.; Mahmud-Un-Nabi, M.A.; Sultana, R.; Tasnim, S.; Noor, F.A.; Sarker, S.K.; Islam, M.T.; et al. Genotypic and phenotypic profiles of antibiotic-resistant bacteria isolated from hospitalised patients in Bangladesh. Trop. Med. Int. Health 2021, 26, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).