Longitudinal Fecal Microbiota Profiles in A Cohort of Non-Hospitalized Adolescents and Young Adults with COVID-19: Associations with SARS-CoV-2 Status and Long-Term Fatigue
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Investigational Program
2.3. Precision Fecal Microbiota Profiling
2.4. Statistical Analyses
2.5. Ethical Considerations
3. Results
3.1. Fecal Bacterial Abundances and SARS-CoV-2 Status at Baseline
3.2. Fecal Bacterial Abundances and SARS-CoV-2 Status at Six Months
3.3. Associations Between Bacterial Taxa at Baseline and Fatigue, PCC and PIFS Among SARS-CoV-2 Positive Participants at Six Months
4. Discussion
4.1. SARS-CoV-2 Status
4.2. SARS-CoV-2 Infection and Severity
4.3. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Behnood, S.A.; Shafran, R.; Bennett, S.D.; Zhang, A.X.; O’Mahoney, L.L.; Stephenson, T.J.; Ladhani, S.N.; De Stavola, B.L.; Viner, R.M.; Swann, O.V. Persistent symptoms following SARS-CoV-2 infection amongst children and young people: A meta-analysis of controlled and uncontrolled studies. J. Infect. 2022, 84, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, R.; Chiappini, E.; Licari, A.; Galli, L.; Marseglia, G.L. Prevalence and clinical presentation of long COVID in children: A systematic review. Eur. J. Pediatr. 2022, 181, 3995–4009. [Google Scholar] [CrossRef] [PubMed]
- Ceban, F.; Ling, S.; Lui, L.M.; Lee, Y.; Gill, H.; Teopiz, K.M.; Rodrigues, N.B.; Subramaniapillai, M.; Di Vincenzo, J.D.; Cao, B.; et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain, Behav. Immun. 2022, 101, 93–135. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.K.; Malone, L.A.; Kokorelis, C.; Petracek, L.S.; Eastin, E.F.; Lobner, K.L.; Neuendorff, L.; Rowe, P.C. Long-Term COVID 19 Sequelae in Adolescents: The Overlap with Orthostatic Intolerance and ME/CFS. Curr. Pediatr. Rep. 2022, 10, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.B.; Murthy, S.; Marshall, J.C.; Relan, P.; Diaz, J.V. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 2022, 22, e102–e107. [Google Scholar] [CrossRef]
- Wong, T.L.; Weitzer, D.J. Long COVID and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)-A Systemic Review and Comparison of Clinical Presentation and Symptomatology. Medicina 2021, 57, 418. [Google Scholar] [CrossRef]
- Hickie, I.; Davenport, T.; Wakefield, D.; Vollmer-Conna, U.; Cameron, B.; Vernon, S.D.; Reeves, W.C.; Lloyd, A. Post-infective and chronic fatigue syndromes precipitated by viral and non-viral pathogens: Prospective cohort study. BMJ 2006, 333, 575. [Google Scholar] [CrossRef]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2022, 20, 323–337. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Silva, M.G.; Falcoff, N.L.; Corradi, G.R.; Di Camillo, N.; Seguel, R.F.; Tabaj, G.C.; Guman, G.R.; de Matteo, E.; Nuñez, M.; Gironacci, M.M. Effect of age on human ACE2 and ACE2-expressing alveolar type II cells levels. Pediatr. Res. 2023, 93, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wu, J.; Zeng, Y.; Li, J.; Yan, J.; Meng, W.; Han, H.; Feng, F.; He, J.; Zhao, S.; et al. SARS-CoV-2 triggered oxidative stress and abnormal energy metabolism in gut microbiota. MedComm 2022, 3, e112. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P. SARS-CoV-2 infections in children: Understanding diverse outcomes. Immunity 2022, 55, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020, 109, 1088–1095. [Google Scholar] [CrossRef]
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef]
- Castagnoli, R.; Votto, M.; Licari, A.; Brambilla, I.; Bruno, R.; Perlini, S.; Rovida, F.; Baldanti, F.; Marseglia, G.L. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Children and Adolescents: A Systematic Review. JAMA Pediatr. 2020, 174, 882–889. [Google Scholar] [CrossRef]
- Ailioaie, L.M.; Ailioaie, C.; Litscher, G. Infection, Dysbiosis and Inflammation Interplay in the COVID Era in Children. Int. J. Mol. Sci. 2023, 24, 10874. [Google Scholar] [CrossRef]
- Villapol, S. Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. J. Lab. Clin. Med. 2020, 226, 57–69. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, W.; Guo, M.; Xiao, C.; Fu, Z.; Yu, S.; Jiang, L.; Wang, S.; Ling, Y.; Liu, F.; et al. Integrated analysis of gut microbiome and host immune responses in COVID-19. Front. Med. 2022, 16, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gu, S.; Chen, Y.; Lu, H.; Shi, D.; Guo, J.; Wu, W.-R.; Yang, Y.; Li, Y.; Xu, K.-J.; et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 2022, 71, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.-Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.-C.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Selvakumar, J.; Havdal, L.B.; Drevvatne, M.; Brodwall, E.M.; Berven, L.L.; Stiansen-Sonerud, T.; Einvik, G.; Leegaard, T.M.; Tjade, T.; Michelsen, A.E.; et al. Prevalence and Characteristics Associated With Post–COVID-19 Condition Among Nonhospitalized Adolescents and Young Adults. JAMA Netw. Open 2023, 6, e235763. [Google Scholar] [CrossRef]
- Berven, L.L.; Selvakumar, J.; Havdal, L.; Stiansen-Sonerud, T.; Einvik, G.; Leegaard, T.M.; Tjade, T.; Michelsen, A.E.; Mollnes, T.E.; Wyller, V.B.B. Inflammatory Markers, Pulmonary Function, and Clinical Symptoms in Acute COVID-19 Among Non-Hospitalized Adolescents and Young Adults. Front. Immunol. 2022, 13, 837288. [Google Scholar]
- Morriss, R.; Wearden, A.; Mullis, R. Exploring the validity of the chalder fatigue scale in chronic fatigue syndrome. J. Psychosom. Res. 1998, 45, 411–417. [Google Scholar] [CrossRef]
- White, P.D.; Goldsmith, K.A.; Johnson, A.L.; Potts, L.; Walwyn, R.; DeCesare, J.C.; Baber, H.L.; Burgess, M.; Clark, L.V.; Cox, D.L.; et al. Comparison of adaptive pacing therapy, cognitive behaviour therapy, graded exercise therapy, and specialist medical care for chronic fatigue syndrome (PACE): A randomised trial. Lancet 2011, 377, 823–836. [Google Scholar] [CrossRef]
- Husakova, M.; Kralik, P.; Babak, V.; Slana, I. Efficiency of DNA Isolation Methods Based on Silica Columns and Magnetic Separation Tested for the Detection of Mycobacterium avium Subsp. Paratuberculosis in Milk and Faeces. Materials 2020, 13, 5112. [Google Scholar] [CrossRef]
- Farup, P.G.; Maseng, M.G. Are Faecal Microbiota Analyses on Species-Level Suitable Clinical Biomarkers? A Pilot Study in Subjects with Morbid Obesity. Microorganisms 2021, 9, 664. [Google Scholar] [CrossRef]
- Derosa, L.; Iebba, V.; Silva, C.A.C.; Piccinno, G.; Wu, G.; Lordello, L.; Routy, B.; Zhao, N.; Thelemaque, C.; Birebent, R.; et al. Custom scoring based on ecological topology of gut microbiota associated with cancer immunotherapy outcome. Cell 2024, 187, 3373–3389.e16. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F.; et al. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front. Cell. Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef] [PubMed]
- Piazzesi, A.; Pane, S.; Del Chierico, F.; Romani, L.; Campana, A.; Palma, P.; Putignani, L. The pediatric gut bacteriome and virome in response to SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2024, 14, 1335450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liang, D.; Li, X.; Liu, J.; Fan, M.; Jing, M.; Wang, Y.; Zhang, Y.; Fang, Y.; Li, D. Characteristics of Gut Microbial Profiles of Offshore Workers and Its Associations With Diet. Front. Nutr. 2022, 9, 904927. [Google Scholar] [CrossRef]
- Romaní-Pérez, M.; López-Almela, I.; Bullich-Vilarrubias, C.; Rueda-Ruzafa, L.; Del Pulgar, E.M.G.; Benítez-Páez, A.; Liebisch, G.; Lamas, J.A.; Sanz, Y. Holdemanella biformis improves glucose tolerance and regulates GLP-1 signaling in obese mice. FASEB J. 2021, 35, e21734. [Google Scholar] [CrossRef]
- García-López, M.; Meier-Kolthoff, J.P.; Tindall, B.J.; Gronow, S.; Woyke, T.; Kyrpides, N.C.; Hahnke, R.L.; Göker, M. Analysis of 1,000 Type-Strain Genomes Improves Taxonomic Classification of Bacteroidetes. Front. Microbiol. 2019, 10, 2083. [Google Scholar] [CrossRef]
- Pudlo, N.A.; Urs, K.; Crawford, R.; Pirani, A.; Atherly, T.; Jimenez, R.; Terrapon, N.; Henrissat, B.; Peterson, D.; Ziemer, C.; et al. Phenotypic and Genomic Diversification in Complex Carbohydrate-Degrading Human Gut Bacteria. mSystems 2022, 7, e0094721. [Google Scholar] [CrossRef]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8, 2345. [Google Scholar] [CrossRef]
- Armstrong, H.; Alipour, M.; Valcheva, R.; Bording-Jorgensen, M.; Jovel, J.; Zaidi, D.; Shah, P.; Lou, Y.; Ebeling, C.; Mason, A.L.; et al. Host immunoglobulin G selectively identifies pathobionts in pediatric inflammatory bowel diseases. Microbiome 2019, 7, 1. [Google Scholar] [CrossRef]
- Hollister, E.B.; Oezguen, N.; Chumpitazi, B.P.; Luna, R.A.; Weidler, E.M.; Rubio-Gonzales, M.; Dahdouli, M.; Cope, J.L.; Mistretta, T.-A.; Raza, S.; et al. Leveraging Human Microbiome Features to Diagnose and Stratify Children with Irritable Bowel Syndrome. J. Mol. Diagn. 2019, 21, 449–461. [Google Scholar] [CrossRef]
- Suskun, C.; Kilic, O.; Yilmaz Ciftdogan, D.; Guven, S.; Karbuz, A.; Ozkaya Parlakay, A.; Kara, Y.; Kacmaz, E.; Sahin, A.; Boga, A.; et al. Intestinal microbiota composition of children with infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and multisystem inflammatory syndrome (MIS-C). Eur. J. Pediatr. 2022, 181, 3175–3191. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch. Dis. Child. 2020, 106, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Richards, E.M.; Handberg, E.M.; Pepine, C.J.; Raizada, M.K. Butyrate Regulates COVID-19-Relevant Genes in Gut Epithelial Organoids From Normotensive Rats. Hypertension 2021, 77, e13–e16. [Google Scholar] [CrossRef] [PubMed]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J.; Barrows, B.D.; Quigley, E.M.; Borody, T.J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, U.; Louis, P.; Goesmann, A.; Henrissat, B.; Duncan, S.H.; Flint, H.J. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ. Microbiol. 2014, 16, 2879–2890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients With COVID-19. Gastroenterology 2022, 162, 548–561.e4. [Google Scholar] [CrossRef]
- Lin, R.; Xiao, M.; Cao, S.; Sun, Y.; Zhao, L.; Mao, X.; Chen, P.; Tong, X.; Ou, Z.; Zhu, H.; et al. Distinct gut microbiota and health outcomes in asymptomatic infection, viral nucleic acid test re-positive, and convalescent COVID-19 cases. mLife 2022, 1, 183–197. [Google Scholar] [CrossRef]
- Farsi, Y.; Tahvildari, A.; Arbabi, M.; Vazife, F.; Sechi, L.A.; Bonjar, A.H.S.; Jamshidi, P.; Nasiri, M.J.; Mirsaeidi, M. Diagnostic, Prognostic, and Therapeutic Roles of Gut Microbiota in COVID-19: A Comprehensive Systematic Review. Front. Cell. Infect. Microbiol. 2022, 12, 804644. [Google Scholar] [CrossRef]
- Schult, D.; Reitmeier, S.; Koyumdzhieva, P.; Lahmer, T.; Middelhoff, M.; Erber, J.; Schneider, J.; Kager, J.; Frolova, M.; Horstmann, J.; et al. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in COVID-19. Gut Microbes. 2022, 14, 2031840. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O. Sutterella Species, IgA-degrading Bacteria in Ulcerative Colitis. Trends Microbiol. 2020, 28, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Squillario, M.; Bonaretti, C.; La Valle, A.; Di Marco, E.; Piccolo, G.; Minuto, N.; Patti, G.; Napoli, F.; Bassi, M.; Maghnie, M.; et al. Gut-microbiota in children and adolescents with obesity: Inferred functional analysis and machine-learning algorithms to classify microorganisms. Sci. Rep. 2023, 13, 11294. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, S.; Zhou, Y.; Disoma, C.; Dong, Z.; Du, A.; Zhang, Y.; Chen, Y.; Huang, W.; Chen, J.; et al. Microbiome Profiling Using Shotgun Metagenomic Sequencing Identified Unique Microorganisms in COVID-19 Patients With Altered Gut Microbiota. Front. Microbiol. 2021, 12, 712081. [Google Scholar] [CrossRef]
- Kim, N.; Gim, J.-A.; Lee, B.J.; Choi, B.I.; Yoon, H.S.; Kim, S.H.; Joo, M.K.; Park, J.-J.; Kim, C. Crosstalk between mucosal microbiota, host gene expression, and sociomedical factors in the progression of colorectal cancer. Sci. Rep. 2022, 12, 13447. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Shang, Y.; Wang, W.; Wu, L.; Han, L.; Wang, Q.; Wang, Z.; Xu, H.; Liu, W. Application of two-dimensional polymerase chain reaction to detect four types of microorganisms in feces for assisted diagnosis of IBD. Clin. Chim. Acta. Int. J. Clin. Chem. 2024, 555, 117802. [Google Scholar] [CrossRef]
- Rocchi, G.; Giovanetti, M.; Benedetti, F.; Borsetti, A.; Ceccarelli, G.; Zella, D.; Altomare, A.; Ciccozzi, M.; Guarino, M.P.L. Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens 2022, 11, 1050. [Google Scholar] [CrossRef]


| Baseline | Six Months Follow-Up | |||
|---|---|---|---|---|
| Characteristics | SARS-CoV-2 Positive (n = 136) | SARS-CoV-2 Negative (n = 32) | SARS-CoV-2 Positive (n = 102) | SARS-CoV-2 Negative (n = 17) |
| Female, n (%) | 78 (57) | 19 (59) | 46 (45) | 12 (71)) |
| Age median [IQR] | 17.0 [14,15,16,17,18,19,20,21] | 16.5 [15,16,17,18,19,20] | 17.0 [14–22.5] | 18.0 [16,17,18,19,20,21] |
| BMI median [IQR] | 22.4 [19.9–25.3] | 21.8 [20.2–24.4] | 22.3 [19.8–25.3] | 21.6 [20–23.8] |
| CFQ fatigue caseness, n (%) | 71 (51) | 14 (45) | 36 (35) | 4 (24) |
| PCC caseness, n (%) | 42 (42) | 7 (41) | ||
| PIFS caseness, n (%) | 12 (12) | 1 (6) | ||
| Bacterial Taxa with Deviating Abundance in SARS-CoV-2 Positive Compared to SARS-CoV-2 Negative | ↑ or ↓ in SARS-CoV-2 Positive at Baseline | ↑ or ↓ in SARS-CoV-2 Positive at Six Months | p-Value |
|---|---|---|---|
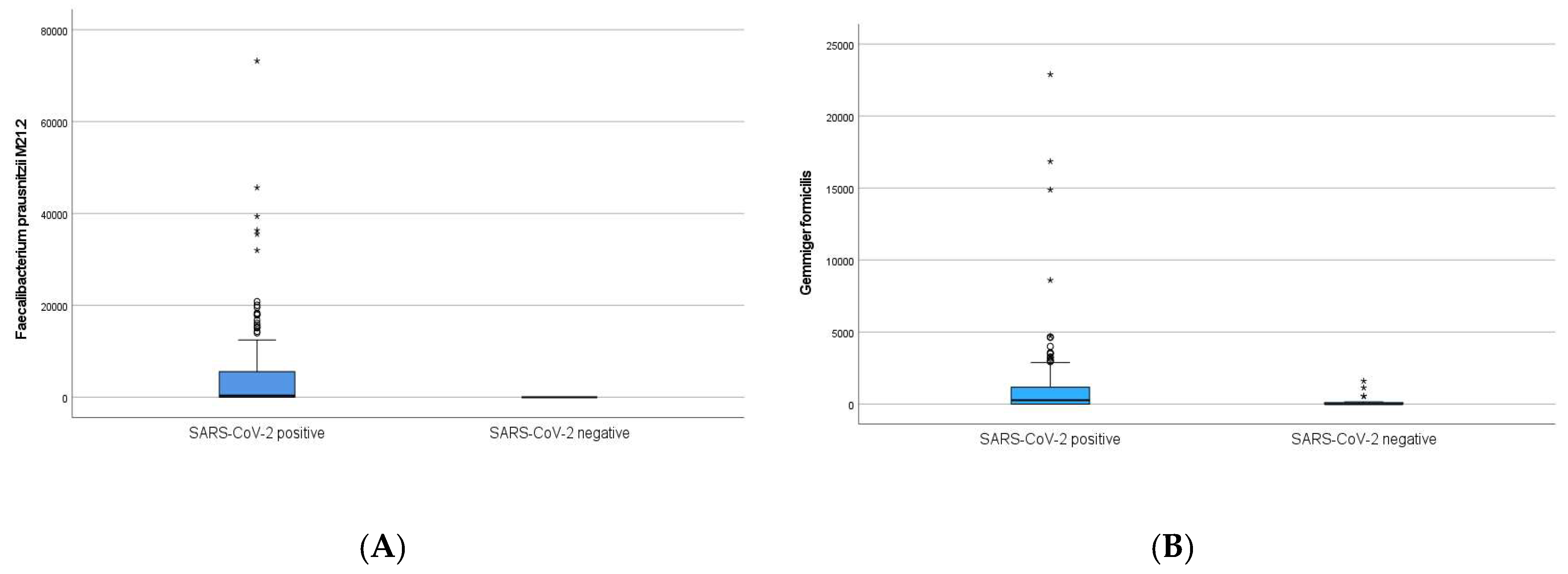
| Faecalibacterium prausnitzii M21.2 | ↑ | ns | <0.001 |
| Gemmiger formicilis | ↑ | ns | <0.001 |
| Gordonibacter pamelaeae | ↑ | ns | 0.003 |
| Holdemanella biformis | ↑ | ns | 0.005 |
| Flavonifractor plautii | ↑ | ns | 0.031 |
| Phocaeicola massiliensis | ↓ | ns | 0.012 |
| Holdemanella filiformis | ↓ | ns | 0.005 |
| Eggerthella lenta | ↑ | ns | 0.021 |
| Odoribacter splanchnicus | ↓ | ns | 0.028 |
| Alistipes shahii | ↓ | ns | 0.032 |
| Alistipes finegoldii | ↓ | ns | 0.035 |
| Bacteroides uniformis | ↓ | ns | 0.035 |
| Clostridium citroniae | ↓ | ns | 0.045 |
| Bifidobacterium longum | ↓ | ns | 0.046 |
| Bifidobacterium animalis | ns | ↓ | 0.007 |
| Faecalibacterium prausnitzii CNCM4575 | ns | ↓ | 0.014 |
| Streptococcus anginosus | ns | ↓ | 0.017 |
| Bacteroides stercoris | ns | ↓ | 0.019 |
| Clostridium nexile | ns | ↓ | 0.038 |
| Parabacterium merdae | ns | ↓ | 0.042 |
| Eubacterium eligens | ns | ↑ | 0.048 |
| Bacterial Taxa with Deviating Abundance in Case Compared to No Case | SARS-CoV-2 Positive Participants (n = 130) | |||
|---|---|---|---|---|
| Baseline ↑ or ↓ (p Value) in Case n (%) | Six-Months Follow-Up ↑ or ↓ (p Value) in Case n (%) | |||
| Fatigue 68 (52%) | Fatigue 46 (53%) | PCC 56 (43%) | PIFS 15 (12%) | |
| Bacteroides thetaiomicron | ↑ (0.026) | ns | ns | ns |
| Sutterella wadsworthensis | ↑ (0.047) | ↑ (<0.047) | ↑ (<0.001) | ns |
| Alistipes putredenis | ↑ (0.049) | ns | ns | ns |
| Bifidobact angulatum | ↓ (0.014) | ns | ns | ns |
| Phocoeicola massiliensis | ↓(0.014) | ns | ns | ns |
| Bacteroides stercoris | ↓ (0.025) | ns | ns | ns |
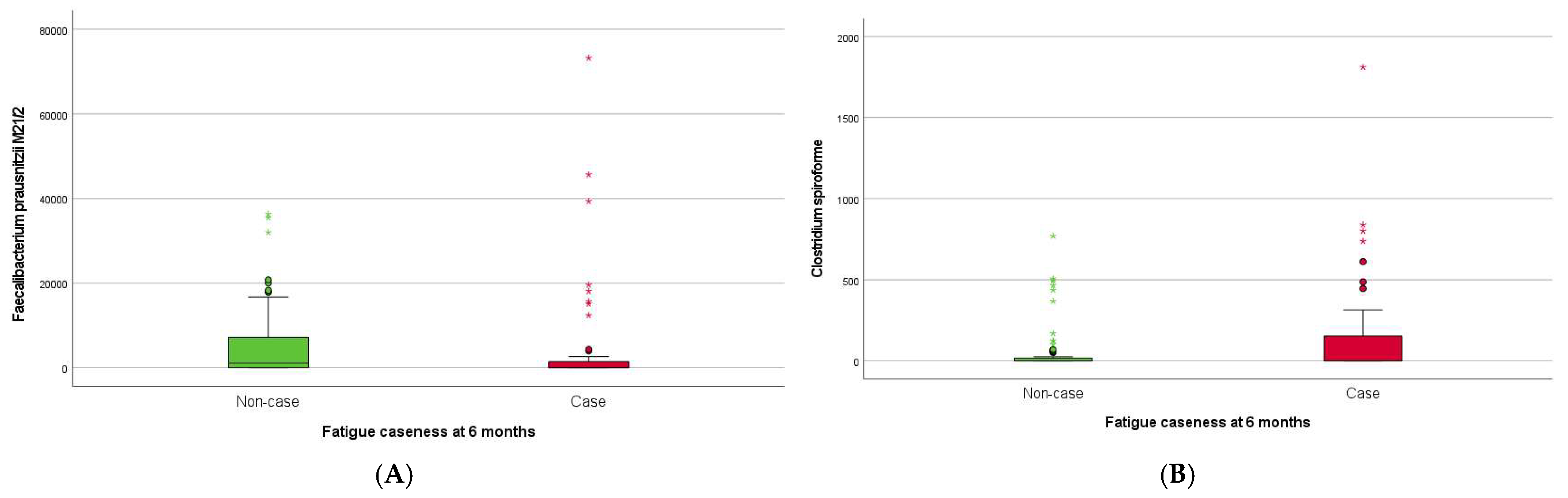
| Clostridium spiroforme | ns | ↑ (0.006) | ns | ns |
| Faecalibacterium prausnitzii CNCM75 | ns | ↑ (0.024) | ns | ns |
| Streptococcus thermophilus | ns | ↑ (0.039) | ↑ (0.042) | ns |
| Roseburia intestinalis | ns | ↑ (0.046) | ns | ns |
| Faecalibacterium prausnitzii M21/2 | ns | ↓ (0.013) | ns | ns |
| Ruminococcus bicirculans | ns | ↓(0.045) | ns | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olbjørn, C.; Hagen, M.; Moen, A.E.F.; Havdal, L.B.; Sommen, S.L.; Berven, L.L.; Thiis-Evensen, E.; Stiansen-Sonerud, T.; Selvakumar, J.; Wyller, V.B.B. Longitudinal Fecal Microbiota Profiles in A Cohort of Non-Hospitalized Adolescents and Young Adults with COVID-19: Associations with SARS-CoV-2 Status and Long-Term Fatigue. Pathogens 2024, 13, 953. https://doi.org/10.3390/pathogens13110953
Olbjørn C, Hagen M, Moen AEF, Havdal LB, Sommen SL, Berven LL, Thiis-Evensen E, Stiansen-Sonerud T, Selvakumar J, Wyller VBB. Longitudinal Fecal Microbiota Profiles in A Cohort of Non-Hospitalized Adolescents and Young Adults with COVID-19: Associations with SARS-CoV-2 Status and Long-Term Fatigue. Pathogens. 2024; 13(11):953. https://doi.org/10.3390/pathogens13110953
Chicago/Turabian StyleOlbjørn, Christine, Milada Hagen, Aina Elisabeth Fossum Moen, Lise Beier Havdal, Silke Lauren Sommen, Lise Lund Berven, Espen Thiis-Evensen, Tonje Stiansen-Sonerud, Joel Selvakumar, and Vegard Bruun Bratholm Wyller. 2024. "Longitudinal Fecal Microbiota Profiles in A Cohort of Non-Hospitalized Adolescents and Young Adults with COVID-19: Associations with SARS-CoV-2 Status and Long-Term Fatigue" Pathogens 13, no. 11: 953. https://doi.org/10.3390/pathogens13110953
APA StyleOlbjørn, C., Hagen, M., Moen, A. E. F., Havdal, L. B., Sommen, S. L., Berven, L. L., Thiis-Evensen, E., Stiansen-Sonerud, T., Selvakumar, J., & Wyller, V. B. B. (2024). Longitudinal Fecal Microbiota Profiles in A Cohort of Non-Hospitalized Adolescents and Young Adults with COVID-19: Associations with SARS-CoV-2 Status and Long-Term Fatigue. Pathogens, 13(11), 953. https://doi.org/10.3390/pathogens13110953






