BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. T. gondii Culture
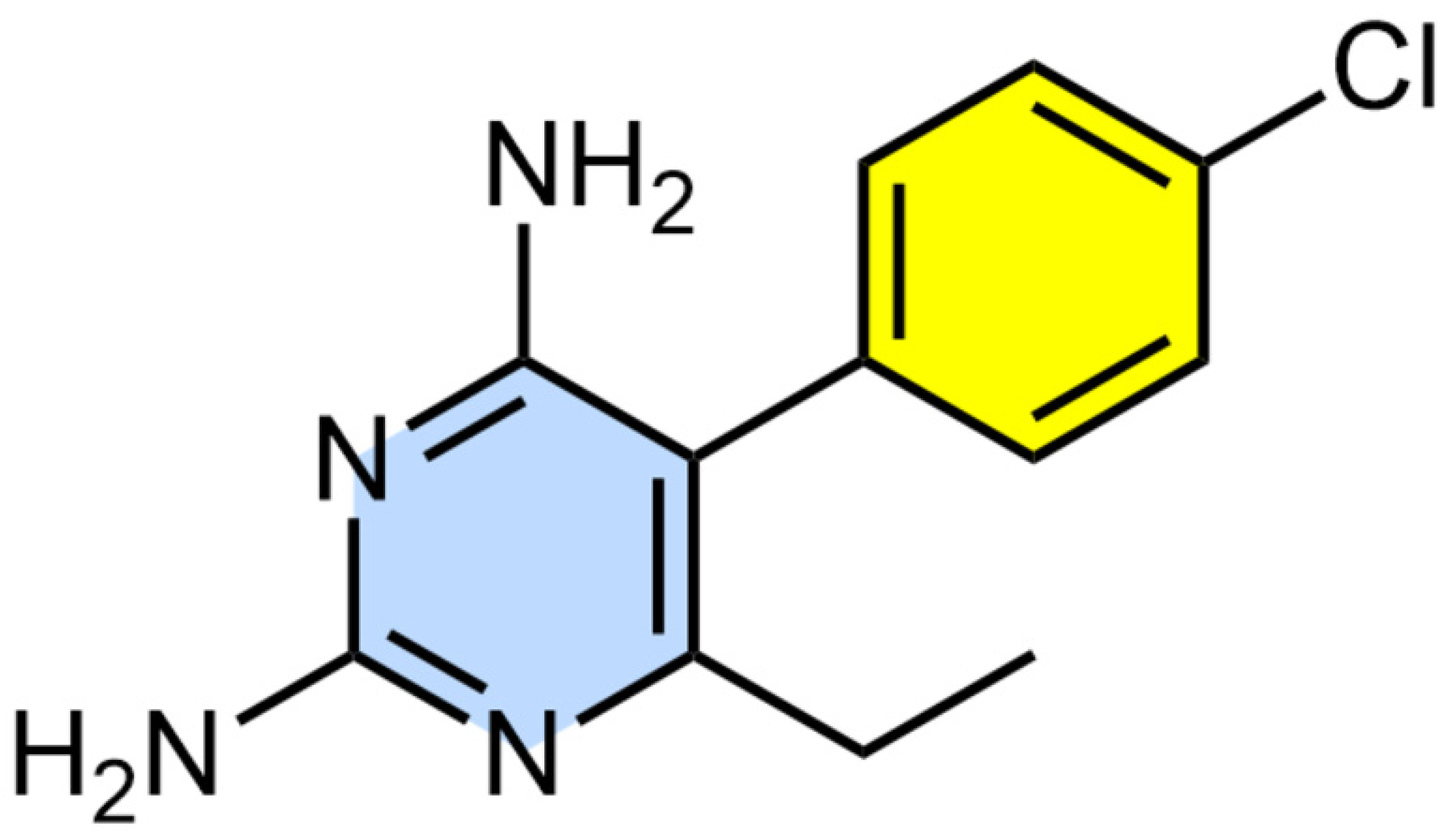
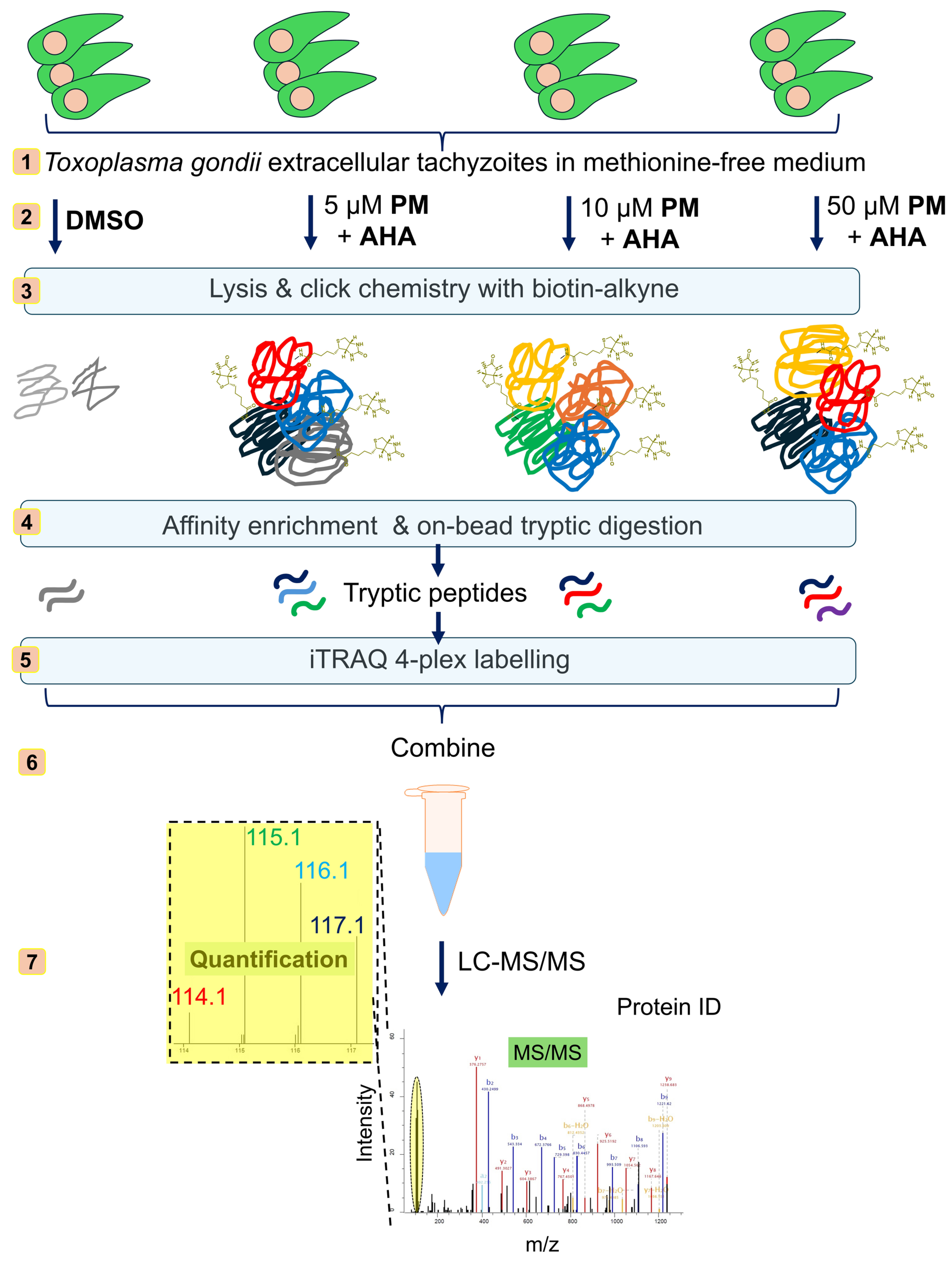
2.2. BONCAT Labelling and PM Treatment
2.3. Click Chemistry-Based Proteome Enrichment, On-Bead Processing and iTRAQ Labelling for Quantitative Analysis
2.4. Liquid Chromatrography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.5. Data Analysis
3. Results
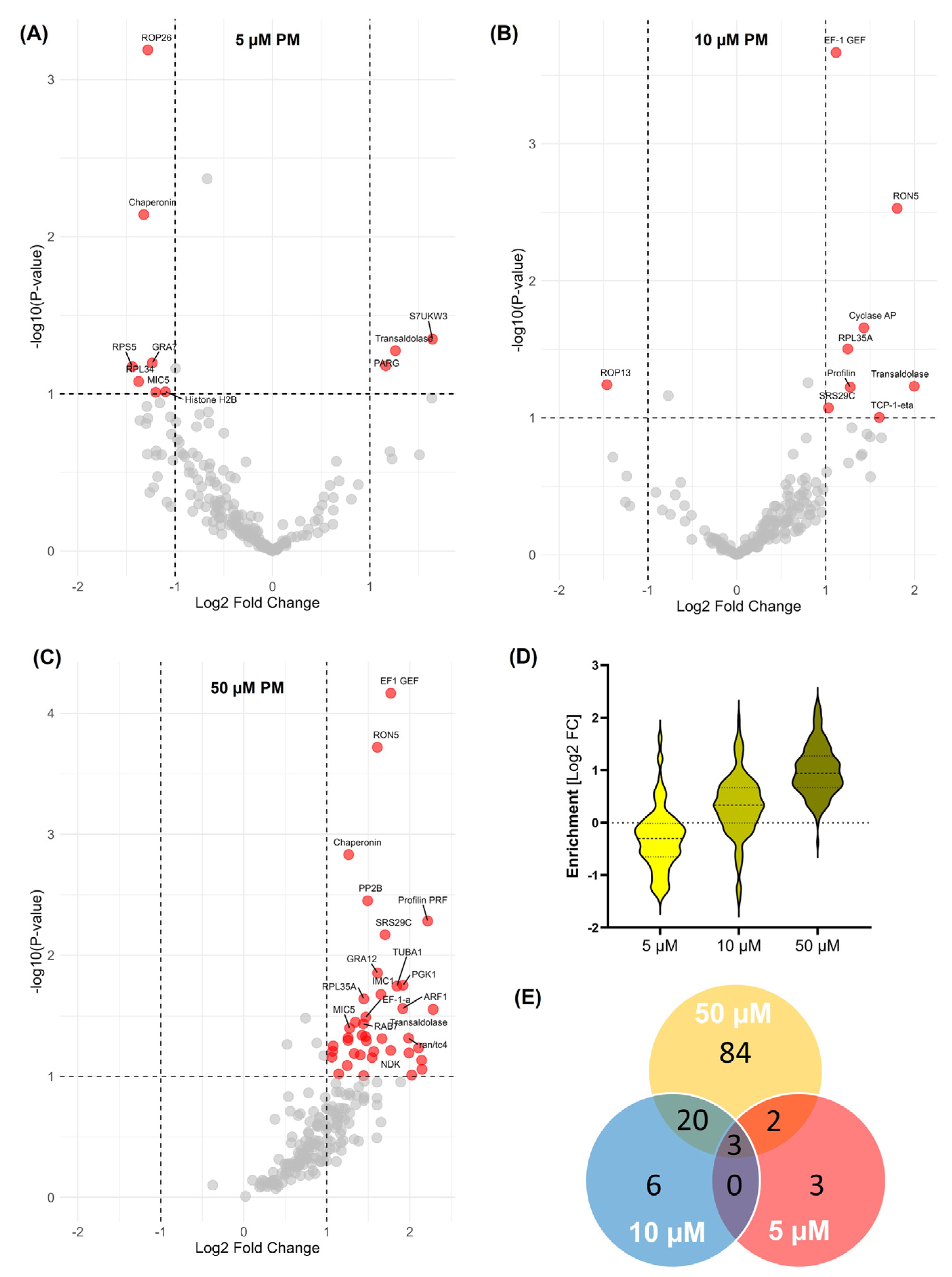
3.1. BONCAT-iTRAQ Profiling of PM-Induced Changes in Global Nascent Protein Synthesis in T. gondii
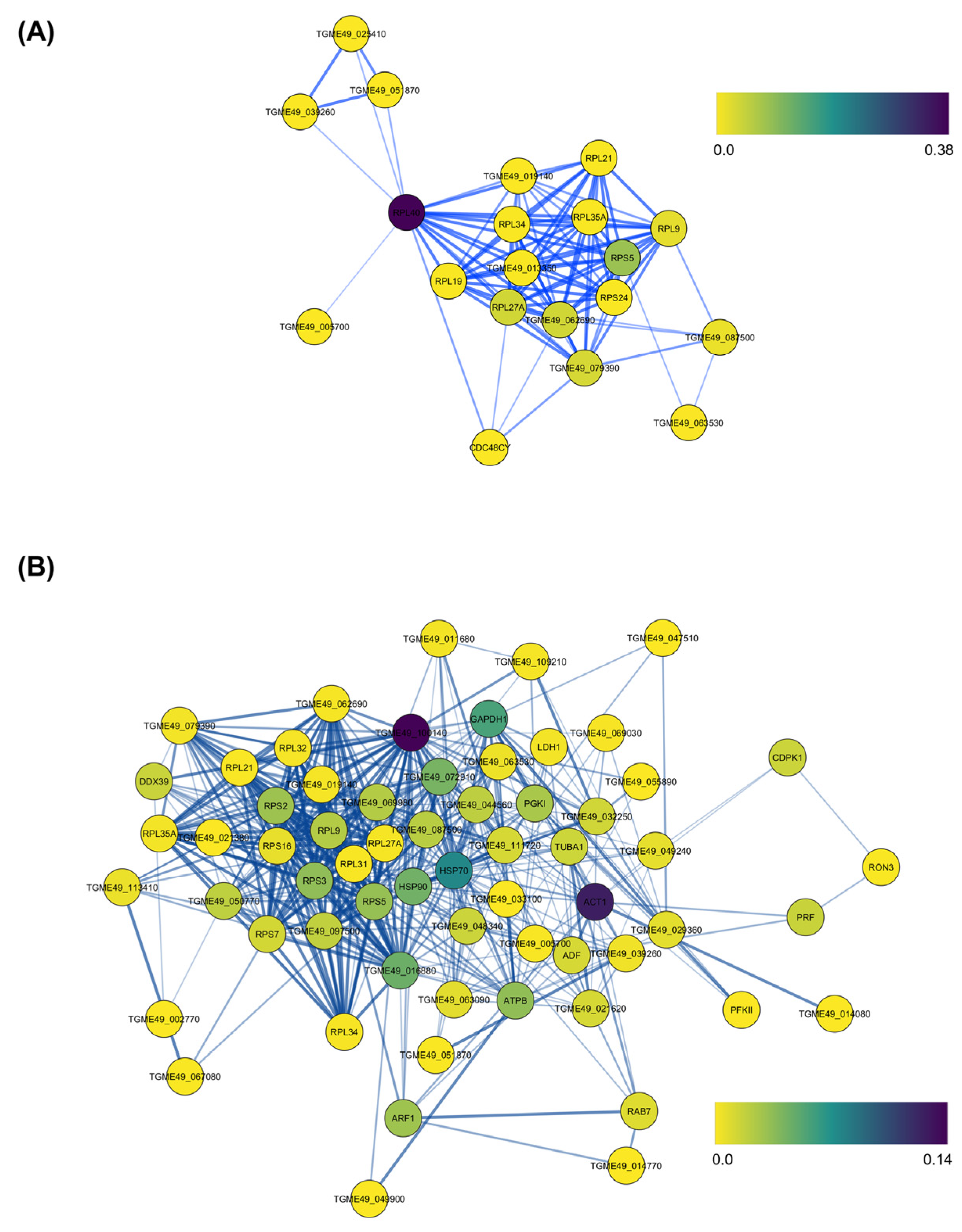
3.2. Protein–Protein Interaction Network Analysis of PM-Modulated NSPs in T. gondii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Heppler, L.N.; Attarha, S.; Persaud, R.; Brown, J.I.; Wang, P.; Petrova, B.; Tosic, I.; Burton, F.B.; Flamand, Y.; Walker, S.R.; et al. The antimicrobial drug pyrimethamine inhibits STAT3 transcriptional activity by targeting the enzyme dihydrofolate reductase. J. Biol. Chem. 2022, 298, 101531. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Oh, J.; Roos, D.S. In vitro generation of novel pyrimethamine resistance mutations in the Toxoplasma gondii dihydrofolate reductase. Antimicrob. Agents Chemother. 2001, 45, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Montazeri, M.; Mehrzadi, S.; Sharif, M.; Sarvi, S.; Tanzifi, A.; Aghayan, S.A.; Daryani, A. Drug Resistance in Toxoplasma gondii. Front. Microbiol. 2018, 9, 2587. [Google Scholar] [CrossRef]
- Garrison, E.M.; Arrizabalaga, G. Disruption of a mitochondrial MutS DNA repair enzyme homologue confers drug resistance in the parasite Toxoplasma gondii. Mol. Microbiol. 2009, 72, 425–441. [Google Scholar] [CrossRef]
- Shen, B.; Powell, R.H.; Behnke, M.S. QTL Mapping and CRISPR/Cas9 Editing to Identify a Drug Resistance Gene in Toxoplasma gondii. J. Vis. Exp. 2017, 124, 55185. [Google Scholar]
- Dieterich, D.C.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA 2006, 103, 9482–9487. [Google Scholar] [CrossRef]
- Dieterich, D.C.; Lee, J.J.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007, 2, 532–540. [Google Scholar] [CrossRef]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [CrossRef]
- Kalesh, K.; Denny, P.W. A BONCAT-iTRAQ method enables temporally resolved quantitative profiling of newly synthesised proteins in Leishmania mexicana parasites during starvation. PLoS Negl. Trop. Dis. 2019, 13, e0007651. [Google Scholar] [CrossRef] [PubMed]
- Kalesh, K.; Sundriyal, S.; Perera, H.; Cobb, S.L.; Denny, P.W. Quantitative Proteomics Reveals that Hsp90 Inhibition Dynamically Regulates Global Protein Synthesis in Leishmania mexicana. mSystems 2021, 6, e00089-21. [Google Scholar] [CrossRef] [PubMed]
- Koutsogiannis, Z.; Mina, J.G.; Albus, C.A.; Kol, M.A.; Holthuis, J.C.M.; Pohl, E.; Denny, P.W. Toxoplasma ceramide synthases: Gene duplication, functional divergence, and roles in parasite fitness. FASEB J. 2023, 37, e23229. [Google Scholar] [CrossRef] [PubMed]
- de Lima Bessa, G.; Vitor, R.W.A.; Lobo, L.M.S.; Rego, W.M.F.; de Souza, G.C.A.; Lopes, R.E.N.; Costa, J.G.L.; Martins-Duarte, E.S. In vitro and in vivo susceptibility to sulfadiazine and pyrimethamine of Toxoplasma gondii strains isolated from Brazilian free wild birds. Sci. Rep. 2023, 13, 7359. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Holmes, M.J.; Shah, P.; Wek, R.C.; Sullivan, W.J., Jr. Simultaneous Ribosome Profiling of Human Host Cells Infected with Toxoplasma gondii. mSphere 2019, 4, e00292-19. [Google Scholar] [CrossRef]
- Wang, X.; Qu, L.; Chen, J.; Jin, Y.; Hu, K.; Zhou, Z.; Zhang, J.; An, Y.; Zheng, J. Toxoplasma rhoptry proteins that affect encephalitis outcome. Cell Death Discov. 2023, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.M.; Xuan, X.; Nishikawa, Y. Toxoplasma gondii cyclophilin 18 regulates the proliferation and migration of murine macrophages and spleen cells. Clin. Vaccine Immunol. 2010, 17, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Narsimulu, B.; Qureshi, R.; Jakkula, P.; Singh, P.; Arifuddin, M.; Qureshi, I.A. Exploration of seryl tRNA synthetase to identify potent inhibitors against leishmanial parasites. Int. J. Biol. Macromol. 2023, 237, 124118. [Google Scholar] [CrossRef]
- Houl, J.H.; Ye, Z.; Brosey, C.A.; Balapiti-Modarage, L.P.F.; Namjoshi, S.; Bacolla, A.; Laverty, D.; Walker, B.L.; Pourfarjam, Y.; Warden, L.S.; et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat. Commun. 2019, 10, 5654. [Google Scholar] [CrossRef] [PubMed]
- Kalesh, K.; Lukauskas, S.; Borg, A.J.; Snijders, A.P.; Ayyappan, V.; Leung, A.K.L.; Haskard, D.O.; DiMaggio, P.A. An Integrated Chemical Proteomics Approach for Quantitative Profiling of Intracellular ADP-Ribosylation. Sci. Rep. 2019, 9, 6655. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Souza, R.O.; Jacobs, K.N.; Back, P.S.; Bradley, P.J.; Arrizabalaga, G. IMC10 and LMF1 mediate mitochondrial morphology through mitochondrion-pellicle contact sites in Toxoplasma gondii. J. Cell Sci. 2022, 135, jcs260083. [Google Scholar] [CrossRef] [PubMed]
- Samland, A.K.; Sprenger, G.A. Transaldolase: From biochemistry to human disease. Int. J. Biochem. Cell Biol. 2009, 41, 1482–1494. [Google Scholar] [CrossRef]
- Guiton, P.S.; Sagawa, J.M.; Fritz, H.M.; Boothroyd, J.C. An in vitro model of intestinal infection reveals a developmentally regulated transcriptome of Toxoplasma sporozoites and a NF-kappaB-like signature in infected host cells. PLoS ONE 2017, 12, e0173018. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Wang, Y.; Gadahi, J.A.; Xu, L.; Yan, R.; Song, X.; Li, X. Toxoplasma gondii Elongation Factor 1-Alpha (TgEF-1alpha) Is a Novel Vaccine Candidate Antigen against Toxoplasmosis. Front. Microbiol. 2017, 8, 168. [Google Scholar]
- Perez, J.M.; Siegal, G.; Kriek, J.; Hard, K.; Dijk, J.; Canters, G.W.; Moller, W. The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli. Structure 1999, 7, 217–226. [Google Scholar] [CrossRef]
- Plattner, F.; Yarovinsky, F.; Romero, S.; Didry, D.; Carlier, M.F.; Sher, A.; Soldati-Favre, D. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 2008, 3, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Tombacz, K.; Burgess, G.; Holder, A.; Werners, A.; Werling, D. Toxoplasma gondii profilin does not stimulate an innate immune response through bovine or human TLR5. Innate Immun. 2018, 24, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.R.; Chen, A.L.; Kim, E.W.; Bradley, P.J. RON5 is critical for organization and function of the Toxoplasma moving junction complex. PLoS Pathog. 2014, 10, e1004025. [Google Scholar] [CrossRef] [PubMed]
- Vaneynde, P.; Verbinnen, I.; Janssens, V. The role of serine/threonine phosphatases in human development: Evidence from congenital disorders. Front. Cell Dev. Biol. 2022, 10, 1030119. [Google Scholar] [CrossRef]
- Hortua Triana, M.A.; Marquez-Nogueras, K.M.; Vella, S.A.; Moreno, S.N.J. Calcium signaling and the lytic cycle of the Apicomplexan parasite Toxoplasma gondii. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1846–1856. [Google Scholar] [CrossRef]
- Rojas-Pirela, M.; Andrade-Alviarez, D.; Rojas, V.; Kemmerling, U.; Caceres, A.J.; Michels, P.A.; Concepcion, J.L.; Quinones, W. Phosphoglycerate kinase: Structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea. Open Biol. 2020, 10, 200302. [Google Scholar] [CrossRef]
- Li, T.; Guo, Y. ADP-Ribosylation Factor Family of Small GTP-Binding Proteins: Their Membrane Recruitment, Activation, Crosstalk and Functions. Front. Cell Dev. Biol. 2022, 10, 813353. [Google Scholar] [CrossRef]
- Liendo, A.; Stedman, T.T.; Ngo, H.M.; Chaturvedi, S.; Hoppe, H.C.; Joiner, K.A. Toxoplasma gondii ADP-ribosylation factor 1 mediates enhanced release of constitutively secreted dense granule proteins. J. Biol. Chem. 2001, 276, 18272–18281. [Google Scholar] [CrossRef]
- Kremer, K.; Kamin, D.; Rittweger, E.; Wilkes, J.; Flammer, H.; Mahler, S.; Heng, J.; Tonkin, C.J.; Langsley, G.; Hell, S.W.; et al. An overexpression screen of Toxoplasma gondii Rab-GTPases reveals distinct transport routes to the micronemes. PLoS Pathog. 2013, 9, e1003213. [Google Scholar] [CrossRef]
- Das, S.; Hehnly, H.; Doxsey, S. A new role for Rab GTPases during early mitotic stages. Small GTPases 2014, 6, 11–15. [Google Scholar] [CrossRef]
- Xue, J.; Jiang, W.; Li, J.; Xiong, W.; Tian, Z.; Zhang, Q.; Li, S.; Liu, C.; Huang, K.; Wang, Q. Toxoplasma gondii RPL40 is a circulating antigen with immune protection effect. Folia Parasitol. 2019, 66, 13. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Mushegian, A.R.; Tatusov, R.L.; Altschul, S.F.; Bryant, S.H.; Bork, P.; Valencia, A. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain--study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994, 3, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.; Alves, C.N.; Lameira, J.; Tunon, I.; Marti, S.; Moliner, V. The catalytic mechanism of glyceraldehyde 3-phosphate dehydrogenase from Trypanosoma cruzi elucidated via the QM/MM approach. Phys. Chem. Chem. Phys. 2013, 15, 3772–3785. [Google Scholar] [CrossRef] [PubMed]
- Kravets, E.; Degrandi, D.; Ma, Q.; Peulen, T.O.; Klumpers, V.; Felekyan, S.; Kuhnemuth, R.; Weidtkamp-Peters, S.; Seidel, C.A.; Pfeffer, K. Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. Elife 2016, 5, e11479. [Google Scholar] [CrossRef]
- Nazarova, L.A.; Ochoa, R.J.; Jones, K.A.; Morrissette, N.S.; Prescher, J.A. Extracellular Toxoplasma gondii tachyzoites metabolize and incorporate unnatural sugars into cellular proteins. Microbes Infect. 2016, 18, 199–210. [Google Scholar] [CrossRef]
- Wang, J.L.; Li, T.T.; Zhang, N.Z.; Wang, M.; Sun, L.X.; Zhang, Z.W.; Fu, B.Q.; Elsheikha, H.M.; Zhu, X.Q. The transcription factor AP2XI-2 is a key negative regulator of Toxoplasma gondii merogony. Nat. Commun. 2024, 15, 793. [Google Scholar] [CrossRef]
| Protein Name | Protein ID | Log2 FC/5 µM | Log2 FC/50 µM |
|---|---|---|---|
| Transaldolase | S7V2K7 | 1.263 | 2.282 |
| Profilin | S7UIQ6 | 0.656 | 2.216 |
| S15 sporozoite-expressed protein | S7UKW3 | 1.643 | 2.147 |
| Proteasome 26S regulatory subunit | S7W860 | N/A | 2.143 |
| Lysine decarboxylase family protein | S7W5X6 | N/A | 2.105 |
| IMC10 | S7WBA5 | 1.229 | 2.024 |
| Fructose-bisphosphatase | A0A125YGG2 | N/A | 1.991 |
| GTP-binding nuclear protein | S7V0L0 | N/A | 1.986 |
| Phosphoglycerate kinase | S7VZ21 | N/A | 1.920 |
| ADP ribosylation factor ARF1 | S7UMN8 | N/A | 1.916 |
| T-complex protein 1 subunit eta | S7UKY6 | N/A | 1.891 |
| Tubulin alpha chain | S7UL74 | N/A | 1.845 |
| EF-1 GEF | S7UX25 | −0.670 | 1.771 |
| PyrDOX | S7VY56 | N/A | 1.771 |
| Major surface antigen | E0AEY9 | −0.499 | 1.702 |
| Heat shock protein HSP70 | A0A125YP12 | N/A | 1.665 |
| IMC1 | A0A125YFV6 | N/A | 1.653 |
| Dense granule protein GRA12 | S7VY87 | N/A | 1.612 |
| Rhoptry neck protein RON5 | S7W8Q7 | −0.274 | 1.610 |
| 26S protease regulatory subunit 4 | A0A125YSZ9 | N/A | 1.606 |
| Preprotein translocase Sec61 | A0A125YSW3 | N/A | 1.606 |
| CDPK1 | S7UHK8 | N/A | 1.606 |
| Putative elongation factor 1-gamma | A0A125YG44 | N/A | 1.566 |
| Nucleoside diphosphate kinase | A0A125YXT7 | N/A | 1.546 |
| Serine/threonine-protein phosphatase | A0A125YWN7 | −0.660 | 1.492 |
| Putative transmembrane protein | S7ULN4 | N/A | 1.478 |
| Elongation factor 1-alpha | S7UIZ8 | −0.771 | 1.470 |
| Rhoptry protein 5B | F2YGR7 | −0.780 | 1.467 |
| Protein disulphide-isomerase | A0A125YQI9 | N/A | 1.453 |
| Putative heat shock protein 90 | S7VTT7 | N/A | 1.451 |
| Prolyl-tRNA synthetase | S7V2A6 | N/A | 1.448 |
| Ribosomal protein RPL35A | A0A125YPQ6 | −0.653 | 1.446 |
| Rhoptry protein ROP15 | S7UKJ6 | N/A | 1.444 |
| GTPase RAB7 | S7V0V9 | N/A | 1.441 |
| Small GTP binding protein rab1a | S7W5S1 | −0.643 | 1.425 |
| DDX39 | A0A125YNR1 | N/A | 1.404 |
| GDPH | S7UQ54 | N/A | 1.386 |
| Actin ACT1 | A0A125YH17 | N/A | 1.360 |
| Cyclophilin | A0A125YTE9 | −0.995 | 1.345 |
| Adenine nucleotide translocator | A0A125YLS5 | N/A | 1.330 |
| RRM-containing protein | A0A125YHM1 | N/A | 1.284 |
| Microneme protein MIC5 | S7W847 | −1.200 | 1.275 |
| Putative chaperonin | S7UXE3 | −1.322 | 1.264 |
| Fructose-bisphosphate aldolase | A0A125YGE5 | −0.903 | 1.258 |
| ATP synthase subunit beta | A0A125YYY4 | N/A | 1.256 |
| Rhoptry protein ROP7 | A0A125YJ16 | N/A | 1.246 |
| Rhoptry kinase family protein ROP26 | S7UKG8 | −1.281 | 1.077 |
| Dense granule protein GRA7 | A0A125YVB8 | −1.235 | 1.071 |
| Ribosomal protein RPL27A | A0A125YTM1 | −1.050 | 1.061 |
| Ribosomal protein RPS5 | A0A125YNF8 | −1.441 | N/A |
| Ribosomal protein RPL34 | A0A125YXF2 | −1.375 | N/A |
| Cell division protein CDC48CY | A0A125YX44 | −1.361 | N/A |
| Rhoptry protein ROP13 | A0A125YQ57 | −1.298 | N/A |
| Ribosomal protein RPL21 | S7WHD1 | −1.291 | N/A |
| Ribosomal protein RPL27 | S7UYE2 | −1.286 | N/A |
| CCT-theta | S7UPG8 | −1.280 | N/A |
| Ribosomal protein RPS15 | A0A125YHZ1 | −1.195 | N/A |
| Ubiquitin | A0A125YNY4 | −1.195 | N/A |
| Ribosomal protein RPL9 | A0A125YRM1 | −1.157 | N/A |
| Seryl-tRNA synthetase | S7WG10 | −1.139 | N/A |
| Histone H2B | A0A125YW36 | −1.100 | N/A |
| Dense granule protein GRA1 | A0A125YRV6 | −1.037 | N/A |
| Rhoptry protein ROP4 | S7UI50 | −1.024 | N/A |
| Rhoptry protein ROP1 | S7W7R3 | −0.992 | N/A |
| Putative peroxiredoxin 6 | S7W128 | −0.976 | N/A |
| 40S ribosomal protein S24 | S7UQS1 | −0.958 | N/A |
| Histone H3 | A0A125YSM0 | −0.943 | N/A |
| Succinate-CoA ligase, beta subunit | A0A125YHZ3 | −0.917 | N/A |
| Histone H4 | A0A125YHU3 | −0.857 | N/A |
| Uncharacterized protein | S7W723 | −0.818 | N/A |
| Uncharacterized protein | A0A125YR22 | −0.750 | N/A |
| 14-3-3 protein | A0A125YLJ3 | −0.705 | N/A |
| Ribosomal protein RPL19 | A0A125YMW0 | −0.651 | N/A |
| Proliferation-associated protein 2G4 | A0A125YVJ6 | −0.504 | N/A |
| Poly(ADP-ribose) glycohydrolase | S7W2K3 | 1.163 | N/A |
| Uncharacterized protein | S7W7E3 | 1.207 | N/A |
| IMP2_N domain-containing protein | S7WAV9 | 1.510 | N/A |
| Acid phosphatase | S7V1S3 | 1.636 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mina, J.G.; Parthasarathy, A.; Porta, E.O.; Denny, P.W.; Kalesh, K. BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment. Pathogens 2024, 13, 879. https://doi.org/10.3390/pathogens13100879
Mina JG, Parthasarathy A, Porta EO, Denny PW, Kalesh K. BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment. Pathogens. 2024; 13(10):879. https://doi.org/10.3390/pathogens13100879
Chicago/Turabian StyleMina, John G., Anutthaman Parthasarathy, Exequiel O. Porta, Paul W. Denny, and Karunakaran Kalesh. 2024. "BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment" Pathogens 13, no. 10: 879. https://doi.org/10.3390/pathogens13100879
APA StyleMina, J. G., Parthasarathy, A., Porta, E. O., Denny, P. W., & Kalesh, K. (2024). BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment. Pathogens, 13(10), 879. https://doi.org/10.3390/pathogens13100879







