A New Approach for Phage Cocktail Design in the Example of Anti-Mastitis Solution
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Collection and Growth Conditions
2.2. Bacteriophages
- —bacteriophage titer (PFU/mL),
- —the sum of plaques on all plates for a given repetition,
- —volume of inoculated phage lysate (0.1 mL),
- —number of plates from 1st dilution,
- —number of plates from 2nd dilution,
- —number of plates from 3rd dilution,
- —the lowest dilution used.
2.3. Characterization of Bacteriophages
2.4. Lytic Activity
2.5. Induction of Phage Resistance
2.6. Bacteriophage Cocktail Host Range (Specificity)
2.7. The Effectiveness of Bacteriophage Cocktail in Prevention of Formation and Eradication of 24 h Bacterial Biofilm
- —effectiveness of biofilm control,
- ATS—absorbance of test sample,
- AC100%—average absorbance of the 100% biofilm control.
2.8. The Effectiveness of Phage Cocktail in the Milk Environment
2.9. Storage Stability of the Bacteriophage Cocktail
3. Results
3.1. Phage Characteristics
3.2. Lytic Activity Results
3.3. Phage Resistance Induced by Selected Phages
3.4. Phage Cocktail Host Range
3.5. The Effectiveness of Bacteriophage Cocktail in Preventing Biofilm Formation and Eradicating 24 h Bacterial Biofilm
3.6. Cocktail Effectiveness in Bacteria Eradication in the Milk Environment
3.7. Storage Stability of Bacteriophage Cocktail
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaczorowski, Ł.; Powierska-Czarny, J.; Wolko, Ł.; Piotrowska-Cyplik, A.; Cyplik, P.; Czarny, J. The Influence of Bacteria Causing Subclinical Mastitis on the Structure of the Cow’s Milk Microbiome. Molecules 2022, 27, 1829. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Gao, Y.; Xue, Y.; Liu, Y.; Zeng, X.; Cheng, Y.; Ma, J.; Wang, H.; Sun, J.; Wang, Z.; et al. Bacteriophage Cocktails Protect Dairy Cows Against Mastitis Caused By Drug Resistant Escherichia Coli Infection. Front. Cell. Infect. Microbiol. 2021, 11, 690377. [Google Scholar] [CrossRef] [PubMed]
- Huma, Z.I.; Sharma, N.; Kour, S.; Lee, S.J. Phenotypic and Molecular Characterization of Bovine Mastitis Milk Origin Bacteria and Linkage of Intramammary Infection with Milk Quality. Front. Vet. Sci. 2022, 9, 885134. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.P.; Ly, S.; Daetwyler, S.; Eichenseher, F.; Loessner, M.J.; Schmelcher, M. Chimeric Peptidoglycan Hydrolases Kill Staphylococcal Mastitis Isolates in Raw Milk and within Bovine Mammary Gland Epithelial Cells. Viruses 2022, 14, 2801. [Google Scholar] [CrossRef]
- Sharun, K.; Dhama, K.; Tiwari, R.; Gugjoo, M.B.; Iqbal Yatoo, M.; Patel, S.K.; Pathak, M.; Karthik, K.; Khurana, S.K.; Singh, R.; et al. Advances in Therapeutic and Managemental Approaches of Bovine Mastitis: A Comprehensive Review. Vet. Q. 2021, 41, 107–136. [Google Scholar] [CrossRef]
- Titze, I.; Lehnherr, T.; Lehnherr, H.; Krömker, V. Efficacy of Bacteriophages against Staphylococcus Aureus Isolates from Bovine Mastitis. Pharmaceuticals 2020, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Saleem, H.D.; Razooqi, M.A.; Gharban, H.A.J. Cumulative Effect of Subclinical Mastitis on Immunological and Biochemical Parameters in Cow Milk. Arch. Razi Inst. 2021, 76, 1629–1638. [Google Scholar] [CrossRef]
- Gomes, F.; Henriques, M. Control of Bovine Mastitis: Old and Recent Therapeutic Approaches. Curr. Microbiol. 2016, 72, 377–382. [Google Scholar] [CrossRef]
- Kovačević, Z.; Samardžija, M.; Horvat, O.; Tomanić, D.; Radinović, M.; Bijelić, K.; Vukomanović, A.G.; Kladar, N. Is There a Relationship between Antimicrobial Use and Antibiotic Resistance of the Most Common Mastitis Pathogens in Dairy Cows? Antibiotics 2023, 12, 3. [Google Scholar] [CrossRef]
- WHO. Available online: https://www.Who.Int/News-Room/Fact-Sheets/Detail/Antibiotic-Resistance (accessed on 14 June 2023).
- WHO. Available online: https://www.Who.Int/News-Room/Fact-Sheets/Detail/Antimicrobial-Resistance (accessed on 14 June 2023).
- Tomanić, D.; Kladar, N.; Radinović, M.; Stančić, I.; Erdeljan, M.; Stanojević, J.; Galić, I.; Bijelić, K.; Kovačević, Z. Intramammary Ethno-Veterinary Formulation in Bovine Mastitis Treatment for Optimization of Antibiotic Use. Pathogens 2023, 12, 259. [Google Scholar] [CrossRef]
- Wang, X.; Li, S.; Du, M.; Liu, N.; Shan, Q.; Zou, Y.; Wang, J.; Zhu, Y. A Novel β-Hairpin Peptide Z-D14CFR Enhances Multidrug-Resistant Bacterial Clearance in a Murine Model of Mastitis. Int. J. Mol. Sci. 2022, 23, 4617. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, Y.D.; Park, J.H.; Moon, G.S. Synergistic Inhibition by Bacteriocin and Bacteriophage against Staphylococcus Aureus. Food Sci. Anim. Resour. 2019, 39, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.; Wong, S.; Jean, J.S.; Slavcev, R. Bacteriophage Interactions with Mammalian Tissue: Therapeutic Applications. Adv. Drug Deliv. Rev. 2019, 145, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Colavecchio, A.; Goodridge, L.D. Phage Therapy Approaches to Reducing Pathogen Persistence and Transmission in Animal Production Environments: Opportunities and Challenges. Microbiol. Spectr. 2017, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, J.; Nakamura, T.; Nakamura, K.; Nishida, K.; Amano, Y.; Watanabe, Y.; Gondaira, S.; Usui, M.; Shimizu, M.; Miyanaga, K.; et al. Biological Properties of Staphylococcus Virus ΦSA012 for Phage Therapy. Sci. Rep. 2022, 12, 21297. [Google Scholar] [CrossRef]
- Kaczorek-Łukowska, E.; Małaczewska, J.; Sowińska, P.; Szymańska, M.; Wójcik, E.A.; Siwicki, A.K. Staphylococcus Aureus from Subclinical Cases of Mastitis in Dairy Cattle in Poland, What Are They Hiding? Antibiotic Resistance and Virulence Profile. Pathogens 2022, 11, 1404. [Google Scholar] [CrossRef]
- Kropinski, A.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 501, ISBN 978-1-58829-682-5. [Google Scholar]
- Su, M.T.; Venkatesh, T.V.; Bodmer, R. Large-and Small-Scale Preparation of Bacterio-Phage λ λ Lysate and DNA; BioTechniques: Huntsville, AL, USA, 1998. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Bilogy 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Cantu, V.A.; Salamon, P.; Seguritan, V.; Redfield, J.; Salamon, D.; Edwards, R.A.; Segall, A.M. PhANNs, a Fast and Accurate Tool and Web Server to Classify Phage Structural Proteins. PLoS Comput. Biol. 2020, 16, e1007845. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-Suite3 for Fast Remote Homology Detection and Deep Protein Annotation. BMC Bioinform. 2019, 20, 473. [Google Scholar] [CrossRef]
- Evans, R.; O’neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2021, 2021, 10. [Google Scholar] [CrossRef]
- Liu, J.; Guo, Z.; Wu, T.; Roy, R.S.; Quadir, F.; Chen, C.; Cheng, J. Enhancing AlphaFold-Multimer-Based Protein Complex Structure Prediction with MULTICOM in CASP15. Commun. Biol. 2023, 6, 1140. [Google Scholar] [CrossRef]
- Zhu, W.; Shenoy, A.; Kundrotas, P.; Elofsson, A. Evaluation of AlphaFold-Multimer Prediction on Multi-Chain Protein Complexes. bioRxiv 2022, 39, btad424. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Seemann, T. Available online: https://Github.Com/Tseemann/Snippy (accessed on 1 January 2020).
- Kutter, E. Phage Host Range and Efficiency of Plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [CrossRef]
- Maszewska, A.; Zygmunt, M.; Grzejdziak, I.; Różalski, A. Use of Polyvalent Bacteriophages to Combat Biofilm of Proteus Mirabilis Causing Catheter-Associated Urinary Tract Infections. J. Appl. Microbiol. 2018, 125, 1253–1265. [Google Scholar] [CrossRef]
- Goulart, D.B.; Mellata, M. Escherichia Coli Mastitis in Dairy Cattle: Etiology, Diagnosis, and Treatment Challenges. Front. Microbiol. 2022, 13, 928346. [Google Scholar] [CrossRef]
- Pascu, C.; Herman, V.; Iancu, I.; Costinar, L. Etiology of Mastitis and Antimicrobial Resistance in Dairy Cattle Farms in the Western Part of Romania. Antibiotics 2022, 11, 57. [Google Scholar] [CrossRef]
- León, M.; Bastías, R. Virulence Reduction in Bacteriophage Resistant Bacteria. Front. Microbiol. 2015, 6, 343. [Google Scholar] [CrossRef]
- Subedi, D.; Gordillo Altamirano, F.; Deehan, R.; Perera, A.; Patwa, R.; Kostoulias, X.; Korneev, D.; Blakeway, L.; Macesic, N.; Peleg, A.Y.; et al. Rational Design of Frontline Institutional Phage Cocktail for the Treatment of Nosocomial Enterobacter Cloacae Complex Infections. bioRxiv 2024. [Google Scholar] [CrossRef]
- Pyzik, E.; Urban-Chmiel, R.; Kurek, Ł.; Herman, K.; Stachura, R.; Marek, A. Bacteriophages for Controlling Staphylococcus Spp. Pathogens on Dairy Cattle Farms: In Vitro Assessment. Animals 2024, 14, 683. [Google Scholar] [CrossRef]
- Rudenko, P.; Sachivkina, N.; Vatnikov, Y.; Shabunin, S.; Engashev, S.; Kontsevaya, S.; Karamyan, A.; Bokov, D.; Kuznetsova, O.; Vasilieva, E. Role of Microorganisms Isolated from Cows with Mastitis in Moscow Region in Biofilm Formation. Vet. World 2021, 14, 40–48. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, Y.; Liu, W.; Zheng, R.; Li, C. Characterization of the Lytic Phage Flora With a Broad Host Range Against Multidrug-Resistant Escherichia Coli and Evaluation of Its Efficacy Against, E. Coli Biofilm Formation. Front. Vet. Sci. 2022, 9, 906973. [Google Scholar] [CrossRef]
- Mohammadian, F.; Rahmani, H.K.; Bidarian, B.; Khoramian, B. Isolation and Evaluation of the Efficacy of Bacteriophages against Multidrug-Resistant (MDR), Methicillin-Resistant (MRSA) and Biofilm-Producing Strains of Staphylococcus Aureus Recovered from Bovine Mastitis. BMC Vet. Res. 2022, 18, 406. [Google Scholar] [CrossRef]
- Teng, F.; Xiong, X.; Zhang, S.; Li, G.; Wang, R.; Zhang, L.; Wang, X.; Zhou, H.; Li, J.; Li, Y.; et al. Efficacy Assessment of Phage Therapy in Treating Staphylococcus Aureus-Induced Mastitis in Mice. Viruses 2022, 14, 620. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Fernández, L.; Martínez, B.; Ruas-Madiedo, P.; García, P.; Rodríguez, A. Real-Time Assessment of Staphylococcus Aureus Biofilm Disruption by Phage-Derived Proteins. Front. Microbiol. 2017, 8, 1632. [Google Scholar] [CrossRef]
- Mi, L.; Liu, Y.; Wang, C.; He, T.; Gao, S.; Xing, S.; Huang, Y.; Fan, H.; Zhang, X.; Yu, W.; et al. Identification of a Lytic Pseudomonas Aeruginosa Phage Depolymerase and Its Anti-Biofilm Effect and Bactericidal Contribution to Serum. Virus Genes. 2019, 55, 394–405. [Google Scholar] [CrossRef]
- Olsen, N.M.C.; Thiran, E.; Hasler, T.; Vanzieleghem, T.; Belibasakis, G.N.; Mahillon, J.; Loessner, M.J.; Schmelcher, M. Synergistic Removal of Static and Dynamic Staphylococcus Aureus Biofilms by Combined Treatment with a Bacteriophage Endolysin and a Polysaccharide Depolymerase. Viruses 2018, 10, 438. [Google Scholar] [CrossRef]
- Guo, Z.; Huang, J.; Yan, G.; Lei, L.; Wang, S.; Yu, L.; Zhou, L.; Gao, A.; Feng, X.; Han, W.; et al. Identification and Characterization of Dpo42, a Novel Depolymerase Derived from the Escherichia Coli Phage VB_EcoM_ECOO78. Front. Microbiol. 2017, 8, 1460. [Google Scholar] [CrossRef]
- Varela-Ortiz, D.F.; Barboza-Corona, J.E.; González-Marrero, J.; León-Galván, M.F.; Valencia-Posadas, M.; Lechuga-Arana, A.A.; Sánchez-Felipe, C.G.; Ledezma-García, F.; Gutiérrez-Chávez, A.J. Antibiotic Susceptibility of Staphylococcus Aureus Isolated from Subclinical Bovine Mastitis Cases and in Vitro Efficacy of Bacteriophage. Vet. Res. Commun. 2018, 42, 243–250. [Google Scholar] [CrossRef]
- Korf, I.H.E.; Kittler, S.; Bierbrodt, A.; Mengden, R.; Rohde, C.; Rohde, M.; Kroj, A.; Lehnherr, T.; Fruth, A.; Flieger, A.; et al. In Vitro Evaluation of a Phage Cocktail Controlling Infections with Escherichia Coli. Viruses 2020, 12, 1470. [Google Scholar] [CrossRef]
- Ngassam-Tchamba, C.; Duprez, J.N.; Fergestad, M.; De Visscher, A.; L’Abee-Lund, T.; De Vliegher, S.; Wasteson, Y.; Touzain, F.; Blanchard, Y.; Lavigne, R.; et al. In Vitro and in Vivo Assessment of Phage Therapy against Staphylococcus Aureus Causing Bovine Mastitis. J. Glob. Antimicrob. Resist. 2020, 22, 762–770. [Google Scholar] [CrossRef]
- Porter, J.; Anderson, J.; Carter, L.; Donjacour, E.; Paros, M. In Vitro Evaluation of a Novel Bacteriophage Cocktail as a Preventative for Bovine Coliform Mastitis. J. Dairy. Sci. 2016, 99, 2053–2062. [Google Scholar] [CrossRef]
- Kaur, G.; Agarwal, R.; Sharma, R.K. Bacteriophage Therapy for Critical and High-Priority Antibiotic-Resistant Bacteria and Phage Cocktail-Antibiotic Formulation Perspective. Food Environ. Virol. 2021, 13, 433–446. [Google Scholar] [CrossRef]
- Preine, F.; Herrera, D.; Scherpenzeel, C.; Kalmus, P.; McCoy, F.; Smulski, S.; Rajala-Schultz, P.; Schmenger, A.; Moroni, P.; Krömker, V. Different European Perspectives on the Treatment of Clinical Mastitis in Lactation. Antibiotics 2022, 11, 1107. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Zou, W.; Zhang, M.; Xu, L.; Liu, F.; Li, X.; Wang, L.; Xu, Y. Evaluation of Phage Therapy in the Treatment of Staphylococcus Aureus-Induced Mastitis in Mice. Folia Microbiol. 2020, 65, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Iwano, H.; Inoue, Y.; Takasago, T.; Kobayashi, H.; Furusawa, T.; Taniguchi, K.; Fujiki, J.; Yokota, H.; Usui, M.; Tanji, Y.; et al. Bacteriophage ΦSA012 Has a Broad Host Range against Staphylococcus Aureus and Effective Lytic Capacity in a Mouse Mastitis Model. Biology 2018, 7, 8. [Google Scholar] [CrossRef]
- García, P.; Madera, C.; Martínez, B.; Rodríguez, A.; Evaristo Suárez, J. Prevalence of Bacteriophages Infecting Staphylococcus Aureus in Dairy Samples and Their Potential as Biocontrol Agents. J. Dairy. Sci. 2009, 92, 3019–3026. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Coffey, A.; Meaney, W.J.; Fitzgerald, G.F.; Ross, R.P. Inhibition of Bacteriophage K Proliferation on Staphylococcus Aureus in Raw Bovine Milk. Lett. Appl. Microbiol. 2005, 41, 274–279. [Google Scholar] [CrossRef]
- Nale, J.Y.; McEwan, N.R. Bacteriophage Therapy to Control Bovine Mastitis: A Review. Antibiotics 2023, 12, 1307. [Google Scholar] [CrossRef]
- Medveďová, A.; Valík, Ľ.; Sirotná, Z.; Liptáková, D. Growth Characterisation of Staphylococcus Aureus in Milk: A Quantitative Approach. Czech J. Food Sci. 2009, 27, 433. [Google Scholar] [CrossRef]
- Peles, F.; Wagner, M.; Varga, L.; Hein, I.; Rieck, P.; Gutser, K.; Keresztúri, P.; Kardos, G.; Turcsányi, I.; Béri, B.; et al. Characterization of Staphylococcus Aureus Strains Isolated from Bovine Milk in Hungary. Int. J. Food Microbiol. 2007, 118, 186–193. [Google Scholar] [CrossRef]
- Nilsson, A.S. Phage Therapy-Constraints and Possibilities. Ups. J. Med. Sci. 2014, 119, 192–198. [Google Scholar] [CrossRef]
- Vasileiou, N.G.C.; Fthenakis, G.C.; Mavrogianni, V.S. Comparison of the Efficacy of Intramammary or Injectable Antibiotic Administration against Staphylococcal Mastitis in Ewes. Pathogens 2022, 11, 1164. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Gilbert, F.B.; Martins, R.P.; Germon, P.; Foucras, G. Progress towards the Elusive Mastitis Vaccines. Vaccines 2022, 10, 296. [Google Scholar] [CrossRef]
- Mansilla, F.; Takagi, M.; Garcia-Castillo, V.; Aso, H.; Nader-Macias, M.E.; Vignolo, G.; Kitazawa, H.; Villena, J. Modulation of Toll-like Receptor-Mediated Innate Immunity in Bovine Intestinal Epithelial Cells by Lactic Acid Bacteria Isolated from Feedlot Cattle. Benef. Microbes 2020, 11, 269–282. [Google Scholar] [CrossRef]
- Peralta, O.A.; Carrasco, C.; Vieytes, C.; Tamayo, M.J.; Muñoz, I.; Sepulveda, S.; Tadich, T.; Duchens, M.; Melendez, P.; Mella, A.; et al. Safety and Efficacy of a Mesenchymal Stem Cell Intramammary Therapy in Dairy Cows with Experimentally Induced Staphylococcus Aureus Clinical Mastitis. Sci. Rep. 2020, 10, 2843. [Google Scholar] [CrossRef]
- Yadav, P.; Yadav, A.B.; Gaur, P.; Mishra, V.; Huma, Z.I.; Sharma, N.; Son, Y.O. Bioengineered Ciprofloxacin-Loaded Chitosan Nanoparticles for the Treatment of Bovine Mastitis. Biomedicines 2022, 10, 3282. [Google Scholar] [CrossRef]







| Bacterial Strain | UWM Strain ID | Year of Isolation |
|---|---|---|
| Escherichia coli 090PP2016 | 407 | 2016 |
| Escherichia coli 091PP2016 | 408 | 2016 |
| Escherichia coli 092PP2016 | 411 | 2016 |
| Escherichia coli 093PP2016 | 412 | 2016 |
| Escherichia coli 094PP2016 | 384 | 2016 |
| Escherichia coli 095PP2016 | 230 | 2016 |
| Escherichia coli 096PP2016 | 375 | 2016 |
| Escherichia coli 097PP2016 | 381 | 2016 |
| Escherichia coli 098PP2016 | 125 | 2016 |
| Escherichia coli 099PP2016 | 513 | 2016 |
| Escherichia coli 100PP2016 | 282 | 2016 |
| Escherichia coli 101PP2016 | 124 | 2016 |
| Escherichia coli 103PP2016 | 419 | 2016 |
| Escherichia coli 104PP2016 | 418 | 2016 |
| Escherichia coli 117PP2016 | 552 | 2016 |
| Escherichia coli 118PP2016 | 551 | 2016 |
| Escherichia coli 132PP2017 | 538 | 2017 |
| Escherichia coli 133PP2017 | 574 | 2017 |
| Staphylococcus aureus 058PP2016 | 377 | 2016 |
| Staphylococcus aureus 059PP2016 | 360 | 2016 |
| Staphylococcus aureus 060PP2016 | 342 | 2016 |
| Staphylococcus aureus 062PP2016 | 312 | 2016 |
| Staphylococcus aureus 063PP2016 | 322 | 2016 |
| Staphylococcus aureus 067PP2016 | 390 | 2016 |
| Staphylococcus aureus 069PP2016 | 522 | 2016 |
| Staphylococcus aureus 075PP2016 | 476 | 2016 |
| Staphylococcus aureus 076PP2016 | 227 | 2016 |
| Staphylococcus aureus 079PP2016 | 294 | 2016 |
| Staphylococcus aureus 080PP2016 | 165 | 2016 |
| Staphylococcus aureus 082PP2016 | 228 | 2016 |
| Staphylococcus aureus 083PP2016 | 536 | 2016 |
| Staphylococcus aureus 090PP2016 | 544 | 2016 |
| Staphylococcus aureus 091PP2016 | 556 | 2016 |
| Bacteriophage | Short Name | Host | Source of Sample |
|---|---|---|---|
| 241Ecol014PP | 241 | E. coli | Water vacuum cleaner |
| 303Ecol101PP | 303 | Waste water 1 | |
| 308Ecol098PP | 308 | Waste water 2 | |
| 310Ecol104PP | 310 | Waste water 1 | |
| 348Ecol098PP | 348 | Waste water 1 | |
| 351Saur083PP | 351 | S. aureus | Waste water 2 |
| 355Saur083PP | 355 | Waste water 2 | |
| 357Saur119PP | 357 | Waste water 2 |
| Feature (GenBank: ID) | 303Ecol101PP (OR062944) | 308Ecol101PP (OR062945) | 310Ecol104PP (OR062946) | 348Ecol098PP (OR062947) | 241Ecol014PP (OR062943) | |
|---|---|---|---|---|---|---|
| Size of genome [bp] | 166,904 | 169,543 | 167,023 | 170,844 | 138,401 | |
| ORF | 265 | 270 | 258 | 265 | 208 | |
| tRNA | 11 | 2 | 10 | 2 | 4 | |
| GC pairs content [%] | 35 | 38 | 36 | 38 | 44 | |
| Taxonomy | Class | Caudoviricetes | Caudoviricetes | Caudoviricetes | Caudoviricetes | Caudoviricetes |
| Family | Straboviridae | Straboviridae | Straboviridae | Straboviridae | - | |
| Subfamily | Tevenvirinae | Tevenvirinae | Tevenvirinae | Tevenvirinae | Vequintavirinae | |
| Genus | Tequatrovirus | Mosigvirus | Tequatrovirus | Mosigvirus | Vequintavirus | |
| Species | Tequatrovirus teqdroes | Mosigvirus mar005p1 | - | - | - | |
| NCBI Reference Sequence: | NC_054932.1 | NC_029091.1 | NC_054932.1 | NC_029091.1 | NC_041869.1 | |
| VIRIDIC [%] | 95.97 | 95.67 | 92.07 | 93.27 | 89.62 | |
| Feature (GenBank: ID) | 351Saur083PP (OR062948) | 355Saur083PP (OR062949) | 357Saur119PP (OR062950) | |
|---|---|---|---|---|
| Size of genome [bp] | 17,209 | 143,709 | 140,580 | |
| ORF | 19 | 216 | 209 | |
| tRNA | 0 | 4 | 4 | |
| GC pairs content [%] | 29 | 30 | 30 | |
| Taxonomy | Class | Caudoviricetes | Caudoviricetes | Caudoviricetes |
| Family | Rountreeviridae | Herelleviridae | Herelleviridae | |
| Subfamily | Rakietenvirinae | Twortvirinae | Twortvirinae | |
| Genus | Rosenblumvirus | Kayvirus | Kayvirus | |
| Species | Rosenblumvirus GRCS | - | - | |
| NCBI Reference Sequence: | NC_023550.1 | NC_005880.2 | NC_005880.2 | |
| VIRIDIC [%] | 95.40 | 91.83 | 90.22 | |
| Bacteriophages | Genus | Receptor Binding Proteins | Predicted Receptor |
|---|---|---|---|
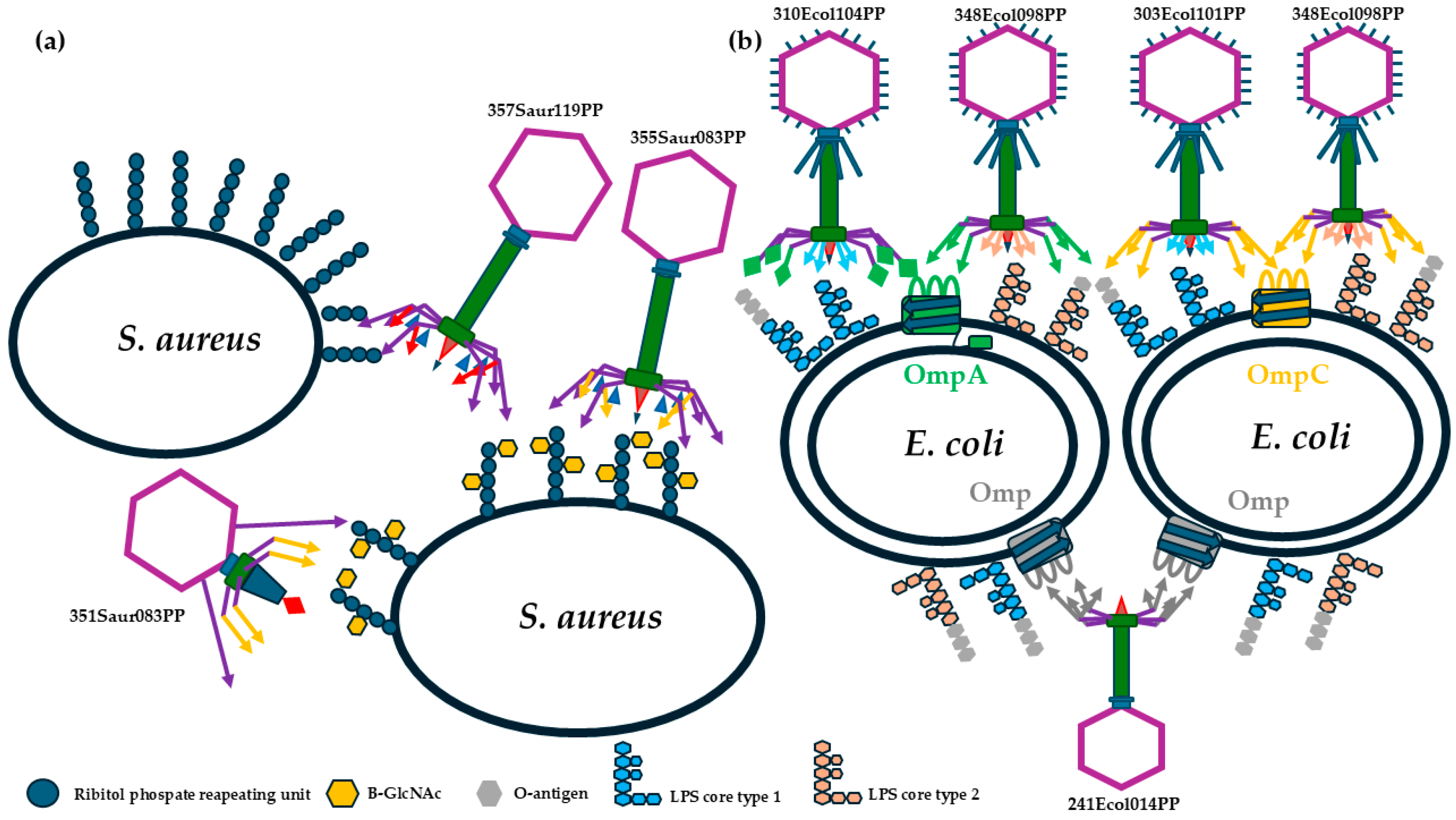
| 351Saur083PP | Rosenblumvirus | WLY86749.1; WLY86757.1; WLY86759.1; WLY86760.1; WLY86762.1 | Main: β-1,4-GlcNAc and cell membrane; Support: unknown oligosaccharide |
| 355Saur083PP | Kayvirus | WLY86864.1; WLY86866.1; WLY86868.1; WLY86875.1 | Main: β-1,4-GlcNAc and cell membrane; Support: unknown oligosaccharide |
| 357Saur119PP | Kayvirus | WLY87089.1; WLY87091.1; WLY87093.1; WLY87100.1 | Main: α-1,4-GlcNAc and cell membrane; Support: unknown oligosaccharide |
| 303Ecol101PP | Tequatrovirus | WLY85731.1; WLY85819.1; WLY85826.1; WLY85828.1 | Main: lipopolysaccharide core the same as phage 310Ecol104PP and OmpC |
| 308Ecol101PP | Mosigvirus | WLY86025.1; WLY86032.1; WLY86034.1; WLY86213.1 | Main: lipopolysaccharide core the same as phage 348Ecol098PP and OmpC |
| 310Ecol104PP | Tequatrovirus | WLY86436.1; WLY86435.1; WLY86348.1; WLY86341.1; WLY86339.1 | Main: lipopolysaccharide core the same as 303Ecol101PP and OmpA and PhoE |
| 348Ecol098PP | Mosigvirus | WLY86505.1; WLY86512.1; WLY86514.1; WLY86683.1 | Main: lipopolysaccharide core the same as phage 308Ecol101PP and OmpC or OmpF or maltoporin |
| 241Ecol014PP | Vequintavirus | WLY85594.1; WLY85596.1; WLY85598.1; WLY85602.1; WLY85606.1; WLY85608.1; WLY85610.1 | Lipopolysaccharide and unknown outer membrane porin |
| % of Strains within the Bacterial Collection | % of Strains within the Bacterial Collection | ||
|---|---|---|---|
| Bacteriophage | Strong Inhibition | Weak Inhibition | No Influence |
| 241Ecol014PP | 33.3% | 22.2% | 44.4% |
| 303Ecol101PP | 27.8% | 22.2% | 50.0% |
| 308Ecol101PP | 44.4% | 5.6% | 50.0% |
| 310Ecol104PP | 44.4% | 11.1% | 44.4% |
| 348Ecol098PP | 50.0% | 5.6% | 44.4% |
| Overlapping activity of all E. coli phages | 83.3% | 0% | 16.7% |
| 351Saur083PP | 40.0% | 20.0% | 40.0% |
| 355Saur083PP | 86.7% | 6.7% | 6.7% |
| 357Saur119PP | 26.7% | 13.3% | 60.0% |
| Overlapping activity of all S. aureus phages | 93.3% | 6.7% | 0% |
| Bacterial Strain Used to Obtain Phage Resistant Variants | Bacteriophage | Obtained Resistant Variant to Specific Bacteriophage | Phage Used in Spot Test | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 303 | 308 | 310 | 348 | 241 | 351 | 355 | 357 | |||
| E. coli 101 | 303 | E. coli 227PP2017 | − | + | + | + | − | ⊗ | ⊗ | ⊗ |
| E. coli 228PP2017 | − | + | + | + | − | ⊗ | ⊗ | ⊗ | ||
| E. coli 229PP2017 | − | + | + | + | − | ⊗ | ⊗ | ⊗ | ||
| E. coli 095 | 303 | E. coli 230PP2017 | − | − | + | + | + | ⊗ | ⊗ | ⊗ |
| E. coli 1304PP2022 | − | − | + | + | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 1306PP2022 | − | − | + | + | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 095 | 308 | E. coli 235PP2017 | + | − | + | + | + | ⊗ | ⊗ | ⊗ |
| E. coli 236PP2017 | + | − | + | + | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 237 PP2017 | + | − | + | + | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 098 | 348 | E. coli 241PP2017 | − | + | + | − | + | ⊗ | ⊗ | ⊗ |
| E. coli 242PP2017 | − | − | − | − | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 243PP2017 | − | − | − | − | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 244PP2017 | − | − | − | − | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 242 | 241 | E. coli 261PP2018 | − | + | − | + | − | ⊗ | ⊗ | ⊗ |
| E. coli 262PP2018 | − | + | − | − | − | ⊗ | ⊗ | ⊗ | ||
| E. coli 263PP2018 | − | + | − | − | − | ⊗ | ⊗ | ⊗ | ||
| E. coli 264PP2018 | − | − | − | − | − | ⊗ | ⊗ | ⊗ | ||
| E. coli 104 | 310 | E. coli 265PP2018 | − | + | − | − | + | ⊗ | ⊗ | ⊗ |
| E. coli 266PP2018 | − | + | − | − | + | ⊗ | ⊗ | ⊗ | ||
| E. coli 300PP2018 | − | + | − | − | + | ⊗ | ⊗ | ⊗ | ||
| S. aureus 083 | 351 | S. aureus 119PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + |
| S. aureus 120PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| S. aureus 121PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| S. aureus 122PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| 355 | S. aureus 123PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | |
| S. aureus 124PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| S. aureus 125PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| S. aureus 126PP2018 | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | − | − | + | ||
| 357 | resistant variants not obtained | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | ⊗ | |
| Bacterial Mutants | NT_POS | AA_POS | Mutation Effect | Gene | Product |
|---|---|---|---|---|---|
| 119PP2018 | 1177/1722 | 393/573 | Stop gained Arg393 * | tarS | Poly(ribitol-phosphate) β-N-acetylglucosaminyltransferase TarS |
| - | - | Intergenic region C1550T | 5SrRNA | 5S ribosomal RNA (partial) | |
| 120PP2018 | 1027/1722 | 343/573 | Stop gained G1027T Glu343 * | tarS | Poly(ribitol-phosphate) β-N-acetylglucosaminyltransferase TarS |
| - | - | Intergenic region C1550T | 5SrRNA | 5S ribosomal RNA (partial) | |
| 121PP2018 | 379/1722 | 127/573 | Missense variant C379T Arg127Cys | tarS | Poly(ribitol-phosphate) β-N-acetylglucosaminyltransferase TarS |
| - | - | Intergenic region C1550T | 5SrRNA | 5S ribosomal RNA (partial) |
| Bacterial Mutants | NT_POS | AA_POS | Mutation Effect | Gene | Product |
|---|---|---|---|---|---|
| 235PP2017 | 603/1104 | 201/367 | Frameshift variant 603–606 del CACT Thr202fs | ompC | outer membrane porin C |
| 1304PP2022 | 447/1104 | 149/367 | Frameshift variant 447–454 del CGCGACCT Phe149fs | ompC | outer membrane porin C |
| 527/978 | 176/325 | Frameshift variant 527–537 del AAAACTTGCAG Lys176f | wzzB | regulator of length of O-antigen component of lipopolysaccharide chains | |
| 1306PP2022 | 511/1104 | 171/367 | Stop gained C511T Gln171 * | ompC | outer membrane porin C |
| 453/978 | 151/325 | Stop gained T453A Tyr151 * | wzzB | regulator of length of O-antigen component of lipopolysaccharide chains | |
| 265PP2018 | 97/1041 | 66/346 | Frameshift variant 193–196 dup CAGG Val66fs | ompA | outer membrane protein 3a (II *;G;d) |
| 303/789 | 101/262 | Frameshift variant 303 del A Lys101fs | wbbD | UDP-Gal:alpha-D-GlcNAc-diphosphoundecaprenol beta-1,3-galactosyltransferase |
| Time [Month] | Titer for Each Component of the Cocktail [PFU/mL] | Final Titer [PFU/mL] | Stability [% Log] | |
|---|---|---|---|---|
| 0 | E. coli | 2.16 × 108 | 3.44 × 108 | 100 |
| S. aureus | 1.28 × 108 | |||
| 3 | E. coli | 1.22 × 108 | 1.90 × 108 | 97 |
| S. aureus | 6.82 × 107 | |||
| 6 | E. coli | 1.25 × 108 | 1.98 × 108 | 97 |
| S. aureus | 7.31 × 107 | |||
| 9 | E. coli | 1.47 × 108 | 2.30 × 108 | 98 |
| S. aureus | 8.30 × 107 | |||
| 12 | E. coli | 9.16 × 107 | 1.36 × 108 | 95 |
| S. aureus | 4.42 × 107 | |||
| 15 | E. coli | 7.26 × 107 | 1.02 × 108 | 94 |
| S. aureus | 2.97 × 107 | |||
| 18 | E. coli | 1.12 × 108 | 1.46 × 108 | 96 |
| S. aureus | 3.43 × 107 | |||
| 21 | E. coli | 1.04 × 108 | 1.35 × 108 | 95 |
| S. aureus | 3.09 × 107 | |||
| 24 | E. coli | 1.04 × 108 | 1.38 × 108 | 95 |
| S. aureus | 3.42 × 107 | |||
| Bacteriophage | Phage Genus | Receptor Predicted from Phage Genome | Receptor Prediction Confirmation by Phage-Resistant Mutants (Mutated Genes) |
|---|---|---|---|
| 303Ecol101PP | Tequatrovirus | LPS core and OmpC | 235PP2017, 1304PP2022, 1306PP2022 (ompC, wzzB), |
| 310Ecol104PP | LPS core and OmpA and PhoE | 265PP2018 (ompA, wbbD) | |
| 308Ecol101PP | Mosigvirus | LPS core and OmpC | 235PP2017 (ompC, wzzB) |
| 348Ecol098PP | LPS core and OmpC or OmpF or maltoporin | 265PP2018 (ompA, wbbD) | |
| 241Ecol014PP | Vequintavirus | Lipopolysaccharide (sugar) and unknown membrane protein | No mutant sequences for analyses |
| 351Saur083PP | Rosenblumvirus | β-O-N-acetylglucosamine of wall teichoic acid and supporting other polysaccharides | 119PP2018, 120PP2018, 121PP2018 (tarS) |
| 355Saur083PP | Kayvirus | β-O-N-acetylglucosamine of wall teichoic acid and supporting other polysaccharides | 119PP2018, 120PP2018, 121PP2018 (tarS) |
| 357Saur119PP | Putative α-O-N-acetylglucosamine of wall teichoic acid and supporting other polysaccharides | No obtained mutant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Królikowska, D.; Szymańska, M.; Krzyżaniak, M.; Guziński, A.; Matusiak, R.; Kajdanek, A.; Kaczorek-Łukowska, E.; Maszewska, A.; Wójcik, E.A.; Dastych, J. A New Approach for Phage Cocktail Design in the Example of Anti-Mastitis Solution. Pathogens 2024, 13, 839. https://doi.org/10.3390/pathogens13100839
Królikowska D, Szymańska M, Krzyżaniak M, Guziński A, Matusiak R, Kajdanek A, Kaczorek-Łukowska E, Maszewska A, Wójcik EA, Dastych J. A New Approach for Phage Cocktail Design in the Example of Anti-Mastitis Solution. Pathogens. 2024; 13(10):839. https://doi.org/10.3390/pathogens13100839
Chicago/Turabian StyleKrólikowska, Daria, Marta Szymańska, Marta Krzyżaniak, Arkadiusz Guziński, Rafał Matusiak, Agnieszka Kajdanek, Edyta Kaczorek-Łukowska, Agnieszka Maszewska, Ewelina A. Wójcik, and Jarosław Dastych. 2024. "A New Approach for Phage Cocktail Design in the Example of Anti-Mastitis Solution" Pathogens 13, no. 10: 839. https://doi.org/10.3390/pathogens13100839
APA StyleKrólikowska, D., Szymańska, M., Krzyżaniak, M., Guziński, A., Matusiak, R., Kajdanek, A., Kaczorek-Łukowska, E., Maszewska, A., Wójcik, E. A., & Dastych, J. (2024). A New Approach for Phage Cocktail Design in the Example of Anti-Mastitis Solution. Pathogens, 13(10), 839. https://doi.org/10.3390/pathogens13100839






