The Role of the NRF2 Pathway in the Pathogenesis of Viral Respiratory Infections
Abstract
1. Introduction
2. The NRF2 Pathway Regulates Cellular Responses to Stress
3. Protective Role of NRF2 in Viral Replication and Pathogenesis
3.1. NRF2 Pathway Activation Prevents Viral Entry
3.2. NRF2 Pathway Activation Hampers Cytosolic Viral Protein Synthesis
3.3. NRF2 Activation-Induced Upregulation of HO-1 Inhibits Viral Replication
3.4. NRF2 Pathway and Interferon Antiviral Responses during Infection with Respiratory Viruses
3.5. Cytoplasmic Autophagy Machinery and NRF2 Pathway
3.6. NRF2-Induced Inhibition of Viral Replication Entails Further Biological Mechanisms such as Sirtuins
4. Respiratory Viruses Leverage or Inhibit NRF2 Activation to Enhance Replication
5. NRF2 Pathway Contribution in Viral Pathogenesis in Cell and Tissue Damage
5.1. Virus-Induced Apoptosis and the NRF2 Pathway
5.2. Ferroptosis and the NRF2 Pathway
6. Therapeutic Potential of NRF2 Activation
7. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clementi, N.; Ghosh, S.; De Santis, M.; Castelli, M.; Criscuolo, E.; Zanoni, I.; Clementi, M.; Mancini, N. Viral Respiratory Pathogens and Lung Injury. Clin. Microbiol. Rev. 2021, 34, e00103-20. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.H.; Cardani, A.; Braciale, T.J. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 2016, 38, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Arnott, A.; Semogas, I.; Falsey, A.R.; Openshaw, P.; Wedzicha, J.A.; Campbell, H.; Nair, H.; Investigators, R. The Etiological Role of Common Respiratory Viruses in Acute Respiratory Infections in Older Adults: A Systematic Review and Meta-analysis. J. Infect. Dis. 2020, 222, S563–S569. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Prim. 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Gain, C.; Song, S.; Angtuaco, T.; Satta, S.; Kelesidis, T. The role of oxidative stress in the pathogenesis of infections with coronaviruses. Front. Microbiol. 2022, 13, 1111930. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Solis, G.; Dela Torre-Aguilar, M.J.; Torres-Borrego, J.; Llorente-Cantarero, F.J.; Fernandez-Gutierrez, F.; Gil-Campos, M.; Tunez-Finana, I.; Perez-Navero, J.L. Oxidative stress and inflamatory plasma biomarkers in respiratory syncytial virus bronchiolitis. Clin. Respir. J. 2017, 11, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. p62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef]
- Lane, D.J.R.; Metselaar, B.; Greenough, M.; Bush, A.I.; Ayton, S.J. Ferroptosis and NRF2: An emerging battlefield in the neurodegeneration of Alzheimer’s disease. Essays Biochem. 2021, 65, 925–940. [Google Scholar] [CrossRef]
- Mondal, A.; Dawson, A.R.; Potts, G.K.; Freiberger, E.C.; Baker, S.F.; Moser, L.A.; Bernard, K.A.; Coon, J.J.; Mehle, A. Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. eLife 2017, 6, e26910. [Google Scholar] [CrossRef]
- Kesic, M.J.; Simmons, S.O.; Bauer, R.; Jaspers, I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Radic. Biol. Med. 2011, 51, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, B.; Messier, E.M.; Janssen, W.J.; Nahreini, P.; Wang, J.; Hartshorn, K.L.; Mason, R.J. Nrf2 protects human alveolar epithelial cells against injury induced by influenza A virus. Respir. Res. 2012, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Arakaki, Y.; Esumi, T.; Kohnomi, S.; Yamamoto, C.; Suzuki, Y.; Takahashi, E.; Konishi, S.; Kido, H.; Kuzuhara, T. Bakuchiol Is a Phenolic Isoprenoid with Novel Enantiomer-selective Anti-influenza A Virus Activity Involving Nrf2 Activation. J. Biol. Chem. 2015, 290, 28001–28017. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.L.; Wang, H.Q.; Wu, P.; Hu, J.; Yin, J.Q.; Wu, S.; Ge, M.; Sun, W.F.; Zhao, J.Y.; Aisa, H.A.; et al. Rupestonic acid derivative YZH-106 suppresses influenza virus replication by activation of heme oxygenase-1-mediated interferon response. Free Radic. Biol. Med. 2016, 96, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Yageta, Y.; Ishii, Y.; Morishima, Y.; Masuko, H.; Ano, S.; Yamadori, T.; Itoh, K.; Takeuchi, K.; Yamamoto, M.; Hizawa, N. Role of Nrf2 in host defense against influenza virus in cigarette smoke-exposed mice. J. Virol. 2011, 85, 4679–4690. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Tate, M.D.; MacKenzie-Kludas, C.J.; Pinar, A.; Zeng, W.; Stutz, A.; Latz, E.; Brown, L.E.; Mansell, A. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 2013, 9, e1003392. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Nobs, S.P.; Heer, A.K.; Kurrer, M.; Klinke, G.; van Rooijen, N.; Vogel, J.; Kopf, M. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 2014, 10, e1004053. [Google Scholar] [CrossRef]
- Huang, C.; Feng, F.; Shi, Y.; Li, W.; Wang, Z.; Zhu, Y.; Yuan, S.; Hu, D.; Dai, J.; Jiang, Q.; et al. Protein Kinase C Inhibitors Reduce SARS-CoV-2 Replication in Cultured Cells. Microbiol. Spectr. 2022, 10, e0105622. [Google Scholar] [CrossRef]
- Cuadrado, A.; Pajares, M.; Benito, C.; Jimenez-Villegas, J.; Escoll, M.; Fernandez-Gines, R.; Garcia Yague, A.J.; Lastra, D.; Manda, G.; Rojo, A.I.; et al. Can Activation of NRF2 Be a Strategy against COVID-19? Trends Pharmacol. Sci. 2020, 41, 598–610. [Google Scholar] [CrossRef]
- Zhao, S.; Ghosh, A.; Lo, C.S.; Chenier, I.; Scholey, J.W.; Filep, J.G.; Ingelfinger, J.R.; Zhang, S.L.; Chan, J.S.D. Nrf2 Deficiency Upregulates Intrarenal Angiotensin-Converting Enzyme-2 and Angiotensin 1-7 Receptor Expression and Attenuates Hypertension and Nephropathy in Diabetic Mice. Endocrinology 2018, 159, 836–852. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Haas de Mello, A.; Morris, D.R.; Jones-Hall, Y.L.; Ivanciuc, T.; Sattler, R.A.; Paessler, S.; Menachery, V.D.; Garofalo, R.P.; Casola, A. SARS-CoV-2 Inhibits NRF2-Mediated Antioxidant Responses in Airway Epithelial Cells and in the Lung of a Murine Model of Infection. Microbiol. Spectr. 2023, 11, e0037823. [Google Scholar] [CrossRef] [PubMed]
- Batra, N.; De Souza, C.; Batra, J.; Raetz, A.G.; Yu, A.M. The HMOX1 Pathway as a Promising Target for the Treatment and Prevention of SARS-CoV-2 of 2019 (COVID-19). Int. J. Mol. Sci. 2020, 21, 6412. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.H.; Zhu, Y.; Mao, J.; Du, R.C. T cell immunobiology and cytokine storm of COVID-19. Scand. J. Immunol. 2021, 93, e12989. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.C.; Chang, L.C.; Tseng, J.C.; Chu, C.M.; Liu, Y.C.; Shieh, W.B. Functional haplotypes in the promoter region of transcription factor Nrf2 in chronic obstructive pulmonary disease. Dis. Markers 2010, 28, 185–193. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hosakote, Y.M.; Liu, T.; Castro, S.M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell Mol. Biol. 2009, 41, 348–357. [Google Scholar] [CrossRef]
- Casola, A.; Burger, N.; Liu, T.; Jamaluddin, M.; Brasier, A.R.; Garofalo, R.P. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with respiratory syncytial virus. Role in viral-induced interferon regulatory factor activation. J. Biol. Chem. 2001, 276, 19715–19722. [Google Scholar] [CrossRef]
- Liu, T.; Castro, S.; Brasier, A.R.; Jamaluddin, M.; Garofalo, R.P.; Casola, A. Reactive oxygen species mediate virus-induced STAT activation: Role of tyrosine phosphatases. J. Biol. Chem. 2004, 279, 2461–2469. [Google Scholar] [CrossRef]
- Komaravelli, N.; Ansar, M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein—Ring finger protein 4 dependent pathway. Free Radic. Biol. Med. 2017, 113, 494–504. [Google Scholar] [CrossRef]
- Ren, K.; Lv, Y.; Zhuo, Y.; Chen, C.; Shi, H.; Guo, L.; Yang, G.; Hou, Y.; Tan, R.X.; Li, E. Suppression of IRG-1 Reduces Inflammatory Cell Infiltration and Lung Injury in Respiratory Syncytial Virus Infection by Reducing Production of Reactive Oxygen Species. J. Virol. 2016, 90, 7313–7322. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.H.; Higgins, M.; Hams, E.; et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef] [PubMed]
- San-Juan-Vergara, H.; Peeples, M.E.; Lockey, R.F.; Mohapatra, S.S. Protein kinase C-alpha activity is required for respiratory syncytial virus fusion to human bronchial epithelial cells. J. Virol. 2004, 78, 13717–13726. [Google Scholar] [CrossRef] [PubMed]
- Ivanciuc, T.; Sbrana, E.; Casola, A.; Garofalo, R.P. Protective Role of Nuclear Factor Erythroid 2-Related Factor 2 Against Respiratory Syncytial Virus and Human Metapneumovirus Infections. Front. Immunol. 2018, 9, 854. [Google Scholar] [CrossRef] [PubMed]
- Castro, S.M.; Guerrero-Plata, A.; Suarez-Real, G.; Adegboyega, P.A.; Colasurdo, G.N.; Khan, A.M.; Garofalo, R.P.; Casola, A. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 2006, 174, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.A.; Leon, M.A.; Cespedes, P.F.; Gomez, R.S.; Canedo-Marroquin, G.; Riquelme, S.A.; Salazar-Echegarai, F.J.; Blancou, P.; Simon, T.; Anegon, I.; et al. Heme Oxygenase-1 Modulates Human Respiratory Syncytial Virus Replication and Lung Pathogenesis during Infection. J. Immunol. 2017, 199, 212–223. [Google Scholar] [CrossRef]
- Cho, H.Y.; Imani, F.; Miller-DeGraff, L.; Walters, D.; Melendi, G.A.; Yamamoto, M.; Polack, F.P.; Kleeberger, S.R. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am. J. Respir. Crit. Care Med. 2009, 179, 138–150. [Google Scholar] [CrossRef]
- Segovia, J.; Sabbah, A.; Mgbemena, V.; Tsai, S.Y.; Chang, T.H.; Berton, M.T.; Morris, I.R.; Allen, I.C.; Ting, J.P.; Bose, S. TLR2/MyD88/NF-kappaB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS ONE 2012, 7, e29695. [Google Scholar] [CrossRef]
- Triantafilou, K.; Kar, S.; Vakakis, E.; Kotecha, S.; Triantafilou, M. Human respiratory syncytial virus viroporin SH: A viral recognition pathway used by the host to signal inflammasome activation. Thorax 2013, 68, 66–75. [Google Scholar] [CrossRef]
- Reed, J.L.; Brewah, Y.A.; Delaney, T.; Welliver, T.; Burwell, T.; Benjamin, E.; Kuta, E.; Kozhich, A.; McKinney, L.; Suzich, J.; et al. Macrophage impairment underlies airway occlusion in primary respiratory syncytial virus bronchiolitis. J. Infect. Dis. 2008, 198, 1783–1793. [Google Scholar] [CrossRef]
- Biagioli, M.C.; Kaul, P.; Singh, I.; Turner, R.B. The role of oxidative stress in rhinovirus induced elaboration of IL-8 by respiratory epithelial cells. Free Radic. Biol. Med. 1999, 26, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; van Dyk, L.F.; Jiang, D.; Dakhama, A.; Li, L.; White, S.R.; Gross, A.; Chu, H.W. Interleukin-1 receptor-associated kinase M (IRAK-M) promotes human rhinovirus infection in lung epithelial cells via the autophagic pathway. Virology 2013, 446, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, V.T.; Kong, Y.; Fedorova, O.; Sharma, L.; Dela Cruz, C.S.; Pyle, A.M.; Iwasaki, A.; Foxman, E.F. Regional Differences in Airway Epithelial Cells Reveal Tradeoff between Defense against Oxidative Stress and Defense against Rhinovirus. Cell Rep. 2018, 24, 3000–3007.e3. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Han, M.S.; Lee, T.H.; Lee, D.B.; Park, J.H.; Lee, S.H.; Kim, T.H. Hydrogen peroxide attenuates rhinovirus-induced anti-viral interferon secretion in sinonasal epithelial cells. Front. Immunol. 2023, 14, 1086381. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, K.; Kar, S.; van Kuppeveld, F.J.; Triantafilou, M. Rhinovirus-induced calcium flux triggers NLRP3 and NLRC5 activation in bronchial cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Ai, F.; Zheng, J.; Zhang, Y.; Fan, T. Inhibition of 12/15-LO ameliorates CVB3-induced myocarditis by activating Nrf2. Chem. Biol. Interact. 2017, 272, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Zhao, X.; Li, C.; Sheng, C.; Li, H. EV71 virus reduces Nrf2 activation to promote production of reactive oxygen species in infected cells. Gut Pathog. 2020, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Tung, W.H.; Hsieh, H.L.; Lee, I.T.; Yang, C.M. Enterovirus 71 induces integrin beta1/EGFR-Rac1-dependent oxidative stress in SK-N-SH cells: Role of HO-1/CO in viral replication. J. Cell. Physiol. 2011, 226, 3316–3329. [Google Scholar] [CrossRef]
- Santangelo, R.; Mancuso, C.; Marchetti, S.; Di Stasio, E.; Pani, G.; Fadda, G. Bilirubin: An Endogenous Molecule with Antiviral Activity in vitro. Front. Pharmacol. 2012, 3, 36. [Google Scholar] [CrossRef][Green Version]
- Lin, T.Y.; Hsia, S.H.; Huang, Y.C.; Wu, C.T.; Chang, L.Y. Proinflammatory cytokine reactions in enterovirus 71 infections of the central nervous system. Clin. Infect. Dis. 2003, 36, 269–274. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, K.; Zang, G.; Chen, T.; Lu, N.; Wang, S.; Zhang, G. Curcumin Inhibits Replication of Human Parainfluenza Virus Type 3 by Affecting Viral Inclusion Body Formation. Biomed. Res. Int. 2021, 2021, 1807293. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Ahmadi, Z.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Curcumin Activates the Nrf2 Pathway and Induces Cellular Protection Against Oxidative Injury. Curr. Mol. Med. 2020, 20, 116–133. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gao, Y.; Ci, X. Role of Nrf2 and Its Activators in Respiratory Diseases. Oxid. Med. Cell. Longev. 2019, 2019, 7090534. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, A.; Nahad, M.P.; Faghihloo, E. The role of Nrf2 transcription factor in viral infection. J. Cell. Biochem. 2018, 119, 6366–6382. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Dinkova-Kostova, A.T. Diffusion dynamics of the Keap1-Cullin3 interaction in single live cells. Biochem. Biophys. Res. Commun. 2013, 433, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Lleres, D.; Swift, S.; Dinkova-Kostova, A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. USA 2013, 110, 15259–15264. [Google Scholar] [CrossRef]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef]
- Iso, T.; Suzuki, T.; Baird, L.; Yamamoto, M. Absolute Amounts and Status of the Nrf2-Keap1-Cul3 Complex within Cells. Mol. Cell. Biol. 2016, 36, 3100–3112. [Google Scholar] [CrossRef]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J.; et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef]
- Kurokawa, H.; Motohashi, H.; Sueno, S.; Kimura, M.; Takagawa, H.; Kanno, Y.; Yamamoto, M.; Tanaka, T. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell. Biol. 2009, 29, 6232–6244. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.K.; McMahon, M.; Plummer, S.M.; Higgins, L.G.; Penning, T.M.; Igarashi, K.; Hayes, J.D. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: Demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis 2009, 30, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; An, C.; Gao, Y.; Leak, R.K.; Chen, J.; Zhang, F. Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog. Neurobiol. 2013, 100, 30–47. [Google Scholar] [CrossRef] [PubMed]
- Jancova, P.; Anzenbacher, P.; Anzenbacherova, E. Phase II drug metabolizing enzymes. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 2010, 154, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jang, J.; Park, S.M.; Yang, S.R. An Update on the Role of Nrf2 in Respiratory Disease: Molecular Mechanisms and Therapeutic Approaches. Int. J. Mol. Sci. 2021, 22, 8406. [Google Scholar] [CrossRef] [PubMed]
- Herengt, A.; Thyrsted, J.; Holm, C.K. NRF2 in Viral Infection. Antioxidants 2021, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Lu, X.; Yin, W.; Fu, H.; Zhang, X.; Cheng, L.; Liu, F.; Jin, C.; Tian, X.; Xie, Y.; et al. Activation of NRF2 blocks HIV replication and apoptosis in macrophages. Heliyon 2023, 9, e12575. [Google Scholar] [CrossRef]
- Staitieh, B.S.; Ding, L.; Neveu, W.A.; Spearman, P.; Guidot, D.M.; Fan, X. HIV-1 decreases Nrf2/ARE activity and phagocytic function in alveolar macrophages. J. Leukoc. Biol. 2017, 102, 517–525. [Google Scholar] [CrossRef]
- Mastrantonio, R.; Cervelli, M.; Pietropaoli, S.; Mariottini, P.; Colasanti, M.; Persichini, T. HIV-Tat Induces the Nrf2/ARE Pathway through NMDA Receptor-Elicited Spermine Oxidase Activation in Human Neuroblastoma Cells. PLoS ONE 2016, 11, e0149802. [Google Scholar] [CrossRef]
- Carvajal-Yepes, M.; Himmelsbach, K.; Schaedler, S.; Ploen, D.; Krause, J.; Ludwig, L.; Weiss, T.; Klingel, K.; Hildt, E. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J. Biol. Chem. 2011, 286, 8941–8951. [Google Scholar] [CrossRef]
- Vomund, S.; Schafer, A.; Parnham, M.J.; Brune, B.; von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.R.; Johnson, B.; Mire, C.E.; Xu, W.; Shabman, R.S.; Speller, L.N.; Leung, D.W.; Geisbert, T.W.; Amarasinghe, G.K.; Basler, C.F. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep. 2014, 6, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Page, A.; Volchkova, V.A.; Reid, S.P.; Mateo, M.; Bagnaud-Baule, A.; Nemirov, K.; Shurtleff, A.C.; Lawrence, P.; Reynard, O.; Ottmann, M.; et al. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep. 2014, 6, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Tuersun, H.; Jiao, S.J.; Zheng, J.H.; Xiao, J.B.; Hasim, A. Functional Role of NRF2 in Cervical Carcinogenesis. PLoS ONE 2015, 10, e0133876. [Google Scholar] [CrossRef]
- Yin, Q.; McBride, J.; Fewell, C.; Lacey, M.; Wang, X.; Lin, Z.; Cameron, J.; Flemington, E.K. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008, 82, 5295–5306. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Gjyshi, O.; Flaherty, S.; Veettil, M.V.; Johnson, K.E.; Chandran, B.; Bottero, V. Kaposi’s sarcoma-associated herpesvirus induces Nrf2 activation in latently infected endothelial cells through SQSTM1 phosphorylation and interaction with polyubiquitinated Keap1. J. Virol. 2015, 89, 2268–2286. [Google Scholar] [CrossRef]
- Gjyshi, O.; Roy, A.; Dutta, S.; Veettil, M.V.; Dutta, D.; Chandran, B. Activated Nrf2 Interacts with Kaposi’s Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression. J. Virol. 2015, 89, 7874–7892. [Google Scholar] [CrossRef]
- Liu, B.; Fang, M.; He, Z.; Cui, D.; Jia, S.; Lin, X.; Xu, X.; Zhou, T.; Liu, W. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis. 2015, 6, e1980. [Google Scholar] [CrossRef]
- Choi, Y.; Jiang, Z.; Shin, W.J.; Jung, J.U. Severe Fever with Thrombocytopenia Syndrome Virus NSs Interacts with TRIM21 To Activate the p62-Keap1-Nrf2 Pathway. J. Virol. 2020, 94, e01684-19. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.A.; Ivanova, O.N.; Mukhtarov, F.S.; Tunitskaya, V.L.; Jansons, J.; Isaguliants, M.G.; Kochetkov, S.N.; Ivanov, A.V. Analysis of the Domains of Hepatitis C Virus Core and NS5A Proteins that Activate the Nrf2/ARE Cascade. Acta Nat. 2016, 8, 123–127. [Google Scholar] [CrossRef]
- Guo, H.; Zhou, T.; Jiang, D.; Cuconati, A.; Xiao, G.H.; Block, T.M.; Guo, J.T. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J. Virol. 2007, 81, 10072–10080. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/beta-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.L.; Lin, Y.S.; Chen, C.L.; Tsai, T.T.; Tsai, C.C.; Wu, Y.W.; Ou, Y.D.; Chu, Y.Y.; Wang, J.M.; Yu, C.Y.; et al. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF-alpha production by mononuclear phagocytes. Sci. Rep. 2016, 6, 32000. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Koh, K.; Kim, Y.E.; Ahn, J.H.; Kim, S. Upregulation of Nrf2 expression by human cytomegalovirus infection protects host cells from oxidative stress. J. Gen. Virol. 2013, 94, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Gjyshi, O.; Bottero, V.; Veettil, M.V.; Dutta, S.; Singh, V.V.; Chikoti, L.; Chandran, B. Kaposi’s sarcoma-associated herpesvirus induces Nrf2 during de novo infection of endothelial cells to create a microenvironment conducive to infection. PLoS Pathog. 2014, 10, e1004460. [Google Scholar] [CrossRef]
- Yu, J.S.; Chen, W.C.; Tseng, C.K.; Lin, C.K.; Hsu, Y.C.; Chen, Y.H.; Lee, J.C. Sulforaphane Suppresses Hepatitis C Virus Replication by Up-Regulating Heme Oxygenase-1 Expression through PI3K/Nrf2 Pathway. PLoS ONE 2016, 11, e0152236. [Google Scholar] [CrossRef]
- Chen, M.H.; Lee, M.Y.; Chuang, J.J.; Li, Y.Z.; Ning, S.T.; Chen, J.C.; Liu, Y.W. Curcumin inhibits HCV replication by induction of heme oxygenase-1 and suppression of AKT. Int. J. Mol. Med. 2012, 30, 1021–1028. [Google Scholar] [CrossRef]
- Tseng, C.K.; Hsu, S.P.; Lin, C.K.; Wu, Y.H.; Lee, J.C.; Young, K.C. Celastrol inhibits hepatitis C virus replication by upregulating heme oxygenase-1 via the JNK MAPK/Nrf2 pathway in human hepatoma cells. Antivir. Res. 2017, 146, 191–200. [Google Scholar] [CrossRef]
- Zhu, Z.; Mathahs, M.M.; Schmidt, W.N. Restoration of type I interferon expression by heme and related tetrapyrroles through inhibition of NS3/4A protease. J. Infect. Dis. 2013, 208, 1653–1663. [Google Scholar] [CrossRef] [PubMed]
- Fillebeen, C.; Rivas-Estilla, A.M.; Bisaillon, M.; Ponka, P.; Muckenthaler, M.; Hentze, M.W.; Koromilas, A.E.; Pantopoulos, K. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C Virus. J. Biol. Chem. 2005, 280, 9049–9057. [Google Scholar] [CrossRef] [PubMed]
- McPhee, F.; Caldera, P.S.; Bemis, G.W.; McDonagh, A.F.; Kuntz, I.D.; Craik, C.S. Bile pigments as HIV-1 protease inhibitors and their effects on HIV-1 viral maturation and infectivity in vitro. Biochem. J. 1996, 320 Pt 2, 681–686. [Google Scholar] [CrossRef]
- Yamada, Y.; Limmon, G.V.; Zheng, D.; Li, N.; Li, L.; Yin, L.; Chow, V.T.; Chen, J.; Engelward, B.P. Major shifts in the spatio-temporal distribution of lung antioxidant enzymes during influenza pneumonia. PLoS ONE 2012, 7, e31494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, J.; Wang, L.; Aliyari, S.; Cheng, G. SARS-CoV-2 virus NSP14 Impairs NRF2/HMOX1 activation by targeting Sirtuin 1. Cell Mol. Immunol. 2022, 19, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Du, J.; Yang, S.; Zheng, B.; Shen, J.; Huang, J.; Cao, L.; Huang, S.; Liu, X.; Guo, L.; et al. SARS-CoV-2 ORF3a sensitizes cells to ferroptosis via Keap1-NRF2 axis. Redox Biol. 2023, 63, 102752. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, S.; Celesia, A.; D’Anneo, A.; Lauricella, M.; Carlisi, D.; De Blasio, A.; Giuliano, M. The Good and Bad of Nrf2: An Update in Cancer and New Perspectives in COVID-19. Int. J. Mol. Sci. 2021, 22, 7963. [Google Scholar] [CrossRef]
- Koyuncu, E.; Budayeva, H.G.; Miteva, Y.V.; Ricci, D.P.; Silhavy, T.J.; Shenk, T.; Cristea, I.M. Sirtuins are evolutionarily conserved viral restriction factors. mBio 2014, 5, e02249-14. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, X.; Liu, D.; Fan, Z.; Hu, X.; Yan, J.; Wang, M.; Gao, G.F. Autophagy is involved in influenza A virus replication. Autophagy 2009, 5, 321–328. [Google Scholar] [CrossRef]
- Cummins, N.W.; Weaver, E.A.; May, S.M.; Croatt, A.J.; Foreman, O.; Kennedy, R.B.; Poland, G.A.; Barry, M.A.; Nath, K.A.; Badley, A.D. Heme oxygenase-1 regulates the immune response to influenza virus infection and vaccination in aged mice. FASEB J. 2012, 26, 2911–2918. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Han, L.; Guo, L.; Zhong, H.; Wang, J. Hemin ameliorates influenza pneumonia by attenuating lung injury and regulating the immune response. Int. J. Antimicrob. Agents 2017, 49, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.J. Viruses and apoptosis. Int. J. Exp. Pathol. 2001, 82, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Shenk, T.E. Viruses and apoptosis. Curr. Opin. Genet. Dev. 1995, 5, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Roulston, A.; Marcellus, R.C.; Branton, P.E. Viruses and apoptosis. Annu. Rev. Microbiol. 1999, 53, 577–628. [Google Scholar] [CrossRef] [PubMed]
- Everett, H.; McFadden, G. Apoptosis: An innate immune response to virus infection. Trends Microbiol. 1999, 7, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, J.; Zeng, R.; Yang, J.; Liu, J.; Zhang, Z.; Song, X.; Yao, Z.; Ma, C.; Li, W.; et al. Respiratory Syncytial Virus Replication Is Promoted by Autophagy-Mediated Inhibition of Apoptosis. J. Virol. 2018, 92, e02193-17. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.; Niu, J.; Xu, Y.; Ma, L.; Lu, A.; Wang, X.; Qian, Z.; Huang, Z.; Jin, X.; et al. Antiviral effects of ferric ammonium citrate. Cell Discov. 2018, 4, 14. [Google Scholar] [CrossRef]
- Kumar, R.; Nayak, M.; Sahoo, G.C.; Pandey, K.; Sarkar, M.C.; Ansari, Y.; Das, V.N.R.; Topno, R.K.; Bhawna; Madhukar, M.; et al. Iron oxide nanoparticles based antiviral activity of H1N1 influenza A virus. J. Infect. Chemother. 2019, 25, 325–329. [Google Scholar] [CrossRef]
- Nencioni, L.; Iuvara, A.; Aquilano, K.; Ciriolo, M.R.; Cozzolino, F.; Rotilio, G.; Garaci, E.; Palamara, A.T. Influenza A virus replication is dependent on an antioxidant pathway that involves GSH and Bcl-2. FASEB J. 2003, 17, 758–760. [Google Scholar] [CrossRef]
- Alsuwaidi, A.R.; Almarzooqi, S.; Albawardi, A.; Benedict, S.; Kochiyil, J.; Mustafa, F.; Hartwig, S.M.; Varga, S.M.; Souid, A.K. Cellular bioenergetics, caspase activity and glutathione in murine lungs infected with influenza A virus. Virology 2013, 446, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Yan, Y.; Liu, Q.; Li, N.; Sheng, M.; Zhang, L.; Li, X.; Liang, Z.; Huang, F.; Liu, K.; et al. Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture. J. Virol. 2015, 89, 10347–10358. [Google Scholar] [CrossRef] [PubMed]
- Edeas, M.; Saleh, J.; Peyssonnaux, C. Iron: Innocent bystander or vicious culprit in COVID-19 pathogenesis? Int. J. Infect. Dis. 2020, 97, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Cavezzi, A.; Troiani, E.; Corrao, S. COVID-19: Hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020, 10, 1271. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Polonikov, A. Endogenous Deficiency of Glutathione as the Most Likely Cause of Serious Manifestations and Death in COVID-19 Patients. ACS Infect. Dis. 2020, 6, 1558–1562. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Nabar, N.R.; Shi, C.S.; Kamenyeva, O.; Xiao, X.; Hwang, I.Y.; Wang, M.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018, 9, 904. [Google Scholar] [CrossRef] [PubMed]
- Salimi, V.; Ramezani, A.; Mirzaei, H.; Tahamtan, A.; Faghihloo, E.; Rezaei, F.; Naseri, M.; Bont, L.; Mokhtari-Azad, T.; Tavakoli-Yaraki, M. Evaluation of the expression level of 12/15 lipoxygenase and the related inflammatory factors (CCL5, CCL3) in respiratory syncytial virus infection in mice model. Microb. Pathog. 2017, 109, 209–213. [Google Scholar] [CrossRef]
- Lin, T.Y.; Chu, C.; Chiu, C.H. Lactoferrin inhibits enterovirus 71 infection of human embryonal rhabdomyosarcoma cells in vitro. J. Infect. Dis. 2002, 186, 1161–1164. [Google Scholar] [CrossRef]
- Ilback, N.G.; Frisk, P.; Mohamed, N.; Gadhasson, I.L.; Blomberg, J.; Friman, G. Virus induces metal-binding proteins and changed trace element balance in the brain during the course of a common human infection (coxsackievirus B3) in mice. Sci. Total Env. 2007, 381, 88–98. [Google Scholar] [CrossRef]
- Wang, M.P.; Joshua, B.; Jin, N.Y.; Du, S.W.; Li, C. Ferroptosis in viral infection: The unexplored possibility. Acta Pharmacol. Sin. 2022, 43, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Kovac, S.; Angelova, P.R.; Holmstrom, K.M.; Zhang, Y.; Dinkova-Kostova, A.T.; Abramov, A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta 2015, 1850, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Davis Volk, A.P.; Moreland, J.G. ROS-containing endosomal compartments: Implications for signaling. Methods Enzym. 2014, 535, 201–224. [Google Scholar] [CrossRef]
- Marsh, M.; Pelchen-Matthews, A. Endocytosis in viral replication. Traffic 2000, 1, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Vale-Costa, S.; Amorim, M.J. Recycling Endosomes and Viral Infection. Viruses 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Pensabene, K.M.; LaMorte, J.; Allender, A.E.; Wehr, J.; Kaur, P.; Savage, M.; Eggler, A.L. Acute Oxidative Stress Can Paradoxically Suppress Human NRF2 Protein Synthesis by Inhibiting Global Protein Translation. Antioxidants 2023, 12, 1735. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Shcherbik, N. Effects of Oxidative Stress on Protein Translation: Implications for Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 21, 2661. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Denison, M.R. Seeking membranes: Positive-strand RNA virus replication complexes. PLoS Biol. 2008, 6, e270. [Google Scholar] [CrossRef]
- Chistovich, L.A.; Malinnikova, T.G.; Stoliarova, E.I. Perception of single-formant vowels with irregular fluctuation of fundamental period and amplitude of glottal signals. Fiziol. Zhurnal SSSR Im. IM Sechenova 1982, 68, 1330–1336. [Google Scholar]
- Hu, Z.; Zhang, Z.; Doo, E.; Coux, O.; Goldberg, A.L.; Liang, T.J. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 1999, 73, 7231–7240. [Google Scholar] [CrossRef]
- Espinoza, J.A.; Gonzalez, P.A.; Kalergis, A.M. Modulation of Antiviral Immunity by Heme Oxygenase-1. Am. J. Pathol. 2017, 187, 487–493. [Google Scholar] [CrossRef]
- Singh, N.; Ahmad, Z.; Baid, N.; Kumar, A. Host heme oxygenase-1: Friend or foe in tackling pathogens? IUBMB Life 2018, 70, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Wasan, H.; Reeta, K.H. Heme oxygenase-1 modulation: A potential therapeutic target for COVID-19 and associated complications. Free Radic. Biol. Med. 2020, 161, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Hamad, R.S.; Al-Kuraishy, H.M.; Alexiou, A.; Papadakis, M.; Ahmed, E.A.; Saad, H.M.; Batiha, G.E. SARS-CoV-2 infection and dysregulation of nuclear factor erythroid-2-related factor 2 (Nrf2) pathway. Cell Stress. Chaperones 2023, 28, 657–673. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, M.; Yuan, C.; Ma, Z.; Li, W.; Zhang, Y.; Su, L.; Xu, J.; Liu, W. Progress of cGAS-STING signaling in response to SARS-CoV-2 infection. Front. Immunol. 2022, 13, 1010911. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Sir, D.; Ou, J.H. Autophagy in viral replication and pathogenesis. Mol. Cells 2010, 29, 1–7. [Google Scholar] [CrossRef]
- Digaleh, H.; Kiaei, M.; Khodagholi, F. Nrf2 and Nrf1 signaling and ER stress crosstalk: Implication for proteasomal degradation and autophagy. Cell. Mol. Life Sci. 2013, 70, 4681–4694. [Google Scholar] [CrossRef]
- Prentice, E.; Jerome, W.G.; Yoshimori, T.; Mizushima, N.; Denison, M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004, 279, 10136–10141. [Google Scholar] [CrossRef]
- Zhao, Z.; Thackray, L.B.; Miller, B.C.; Lynn, T.M.; Becker, M.M.; Ward, E.; Mizushima, N.N.; Denison, M.R.; Virgin, H.W.t. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 2007, 3, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Hur, E.G.; Ryoo, I.G.; Jung, K.A.; Kwak, J.; Kwak, M.K. Involvement of the Nrf2-proteasome pathway in the endoplasmic reticulum stress response in pancreatic beta-cells. Toxicol. Appl. Pharmacol. 2012, 264, 431–438. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Monastyrska, I.; Verheije, M.H.; Cali, T.; Ulasli, M.; Bianchi, S.; Bernasconi, R.; de Haan, C.A.; Molinari, M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 2010, 7, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.M.; Tsueng, G.; Sin, J.; Mangale, V.; Rahawi, S.; McIntyre, L.L.; Williams, W.; Kha, N.; Cruz, C.; Hancock, B.M.; et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014, 10, e1004045. [Google Scholar] [CrossRef] [PubMed]
- Berryman, S.; Brooks, E.; Burman, A.; Hawes, P.; Roberts, R.; Netherton, C.; Monaghan, P.; Whelband, M.; Cottam, E.; Elazar, Z.; et al. Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J. Virol. 2012, 86, 12940–12953. [Google Scholar] [CrossRef]
- Wong, J.; Zhang, J.; Si, X.; Gao, G.; Mao, I.; McManus, B.M.; Luo, H. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008, 82, 9143–9153. [Google Scholar] [CrossRef]
- O’Donnell, V.; Pacheco, J.M.; LaRocco, M.; Burrage, T.; Jackson, W.; Rodriguez, L.L.; Borca, M.V.; Baxt, B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 2011, 410, 142–150. [Google Scholar] [CrossRef]
- Staring, J.; von Castelmur, E.; Blomen, V.A.; van den Hengel, L.G.; Brockmann, M.; Baggen, J.; Thibaut, H.J.; Nieuwenhuis, J.; Janssen, H.; van Kuppeveld, F.J.; et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 2017, 541, 412–416. [Google Scholar] [CrossRef]
- Shi, J.; Wong, J.; Piesik, P.; Fung, G.; Zhang, J.; Jagdeo, J.; Li, X.; Jan, E.; Luo, H. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 2013, 9, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Zhang, G.; Yang, X.; Zhang, S.; Chen, L.; Yan, Q.; Xu, M.; Banerjee, A.K.; Chen, M. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 2014, 15, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Zhang, L.; Li, Z.; Zhong, Y.; Tang, Q.; Qin, Y.; Chen, M. The Matrix Protein of Human Parainfluenza Virus Type 3 Induces Mitophagy that Suppresses Interferon Responses. Cell Host Microbe 2017, 21, 538–547.e4. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; Mack, N.J.; Ahmad, N. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 2018, 28, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Patel, S.; Majumdar, A. Role of NRF2 and Sirtuin activators in COVID-19. Clin. Immunol. 2021, 233, 108879. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Park, S.H.; Chang, H.C.; Shapiro, J.S.; Vassilopoulos, A.; Sawicki, K.T.; Chen, C.; Shang, M.; Burridge, P.W.; Epting, C.L.; et al. Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J. Clin. Investig. 2017, 127, 1505–1516. [Google Scholar] [CrossRef]
- Li, Q.; He, M.; Zhou, F.; Ye, F.; Gao, S.J. Activation of Kaposi’s sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: Identification of sirtuin 1 as a regulator of the KSHV life cycle. J. Virol. 2014, 88, 6355–6367. [Google Scholar] [CrossRef]
- Campagna, M.; Herranz, D.; Garcia, M.A.; Marcos-Villar, L.; Gonzalez-Santamaria, J.; Gallego, P.; Gutierrez, S.; Collado, M.; Serrano, M.; Esteban, M.; et al. SIRT1 stabilizes PML promoting its sumoylation. Cell Death Differ. 2011, 18, 72–79. [Google Scholar] [CrossRef]
- Pagans, S.; Pedal, A.; North, B.J.; Kaehlcke, K.; Marshall, B.L.; Dorr, A.; Hetzer-Egger, C.; Henklein, P.; Frye, R.; McBurney, M.W.; et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005, 3, e41. [Google Scholar] [CrossRef]
- Kwon, H.S.; Brent, M.M.; Getachew, R.; Jayakumar, P.; Chen, L.F.; Schnolzer, M.; McBurney, M.W.; Marmorstein, R.; Greene, W.C.; Ott, M. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe 2008, 3, 158–167. [Google Scholar] [CrossRef]
- De Angelis, M.; Amatore, D.; Checconi, P.; Zevini, A.; Fraternale, A.; Magnani, M.; Hiscott, J.; De Chiara, G.; Palamara, A.T.; Nencioni, L. Influenza Virus Down-Modulates G6PD Expression and Activity to Induce Oxidative Stress and Promote Its Replication. Front. Cell Infect. Microbiol. 2021, 11, 804976. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Seong, R.K.; Shin, O.S. Enhanced Viral Replication by Cellular Replicative Senescence. Immune Netw. 2016, 16, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Li, T.; Li, J.H.; Miao, S.Y.; Xiao, X.Z. The Effects of Resveratrol on Inflammation and Oxidative Stress in a Rat Model of Chronic Obstructive Pulmonary Disease. Molecules 2017, 22, 1529. [Google Scholar] [CrossRef]
- Li, G.; Xia, Z.; Liu, Y.; Meng, F.; Wu, X.; Fang, Y.; Zhang, C.; Liu, D. SIRT1 inhibits rheumatoid arthritis fibroblast-like synoviocyte aggressiveness and inflammatory response via suppressing NF-kappaB pathway. Biosci. Rep. 2018, 38, BSR20180541. [Google Scholar] [CrossRef]
- Zhou, J.; Xue, Z.; He, H.N.; Liu, X.; Yin, S.Y.; Wu, D.Y.; Zhang, X.; Schatten, H.; Miao, Y.L. Resveratrol delays postovulatory aging of mouse oocytes through activating mitophagy. Aging 2019, 11, 11504–11519. [Google Scholar] [CrossRef]
- Rossi, G.A.; Sacco, O.; Capizzi, A.; Mastromarino, P. Can Resveratrol-Inhaled Formulations Be Considered Potential Adjunct Treatments for COVID-19? Front. Immunol. 2021, 12, 670955. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Hildt, E. Effect of Hepatitis Viruses on the Nrf2/Keap1-Signaling Pathway and Its Impact on Viral Replication and Pathogenesis. Int. J. Mol. Sci. 2019, 20, 4659. [Google Scholar] [CrossRef]
- Komaravelli, N.; Tian, B.; Ivanciuc, T.; Mautemps, N.; Brasier, A.R.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic. Biol. Med. 2015, 88, 391–403. [Google Scholar] [CrossRef]
- Rashid, M.U.; Gao, A.; Coombs, K.M. Influenza A Virus Uses PSMA2 for Downregulation of the NRF2-Mediated Oxidative Stress Response. J. Virol. 2022, 96, e0199021. [Google Scholar] [CrossRef]
- Andrew, L.D.; Joseph, F.; Sarah, E.-Q.; Aurica, G.T.; Patrick, M.; Kazuhiro, I.; Peter, J.B.; Sebastian, J.; Ian, M.A. LSC Abstract—Rhinovirus infection induces NRF2 in monocytes but not in epithelial cells, via distinct intracellular pathways. Eur. Respir. J. 2015, 46, PA2607. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Tan, K.S.; Lim, R.L.; Liu, J.; Ong, H.H.; Tan, V.J.; Lim, H.F.; Chung, K.F.; Adcock, I.M.; Chow, V.T.; Wang, Y. Respiratory Viral Infections in Exacerbation of Chronic Airway Inflammatory Diseases: Novel Mechanisms and Insights From the Upper Airway Epithelium. Front. Cell Dev. Biol. 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, F.; Liu, T.; Chen, F.; Liu, S.; Yang, J. The role of oxidative stress in influenza virus infection. Microbes Infect. 2017, 19, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.K.; Minakuchi, M.; Wuputra, K.; Ku, C.C.; Pan, J.B.; Kuo, K.K.; Lin, Y.C.; Saito, S.; Lin, C.S.; Yokoyama, K.K. Redox control in the pathophysiology of influenza virus infection. BMC Microbiol. 2020, 20, 214. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Kasahara, K. The emerging role of oxidative stress in complications of COVID-19 and potential therapeutic approach to diminish oxidative stress. Respir. Med. 2021, 187, 106605. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Jaiswal, A.K. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J. Biol. Chem. 2012, 287, 9873–9886. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lu, H.; Bai, Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 2019, 8, 2252–2267. [Google Scholar] [CrossRef]
- Elsby, R.; Kitteringham, N.R.; Goldring, C.E.; Lovatt, C.A.; Chamberlain, M.; Henderson, C.J.; Wolf, C.R.; Park, B.K. Increased constitutive c-Jun N-terminal kinase signaling in mice lacking glutathione S-transferase Pi. J. Biol. Chem. 2003, 278, 22243–22249. [Google Scholar] [CrossRef]
- Rushworth, S.A.; MacEwan, D.J. HO-1 underlies resistance of AML cells to TNF-induced apoptosis. Blood 2008, 111, 3793–3801. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Long, D. Nrf2 and Ferroptosis: A New Research Direction for Neurodegenerative Diseases. Front. Neurosci. 2020, 14, 267. [Google Scholar] [CrossRef] [PubMed]
- Tomiotto-Pellissier, F.; Alves, D.R.; Miranda-Sapla, M.M.; de Morais, S.M.; Assolini, J.P.; da Silva Bortoleti, B.T.; Goncalves, M.D.; Cataneo, A.H.D.; Kian, D.; Madeira, T.B.; et al. Caryocar coriaceum extracts exert leishmanicidal effect acting in promastigote forms by apoptosis-like mechanism and intracellular amastigotes by Nrf2/HO-1/ferritin dependent response and iron depletion: Leishmanicidal effect of Caryocar coriaceum leaf exracts. Biomed. Pharmacother. 2018, 98, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Cataneo, A.H.D.; Tomiotto-Pellissier, F.; Miranda-Sapla, M.M.; Assolini, J.P.; Panis, C.; Kian, D.; Yamauchi, L.M.; Colado Simao, A.N.; Casagrande, R.; Pinge-Filho, P.; et al. Quercetin promotes antipromastigote effect by increasing the ROS production and anti-amastigote by upregulating Nrf2/HO-1 expression, affecting iron availability. Biomed. Pharmacother. 2019, 113, 108745. [Google Scholar] [CrossRef] [PubMed]
- Selvakumar, G.P.; Ahmed, M.E.; Raikwar, S.P.; Thangavel, R.; Kempuraj, D.; Dubova, I.; Saeed, D.; Zahoor, H.; Premkumar, K.; Zaheer, S.; et al. CRISPR/Cas9 Editing of Glia Maturation Factor Regulates Mitochondrial Dynamics by Attenuation of the NRF2/HO-1 Dependent Ferritin Activation in Glial Cells. J. Neuroimmune Pharmacol. 2019, 14, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Fu, C.Y.; Ziros, P.G.; Sykiotis, G.P. Patent Review (2017–2020) of the Keap1/Nrf2 Pathway Using PatSeer Pro: Focus on Autoimmune Diseases. Antioxidants 2020, 9, 1138. [Google Scholar] [CrossRef] [PubMed]
- Barati, M.T.; Caster, D.J. The Potential of Nrf2 Activation as a Therapeutic Target in Systemic Lupus Erythematosus. Metabolites 2022, 12, 151. [Google Scholar] [CrossRef]
- Pokharel, S.M.; Shil, N.K.; Bose, S. Autophagy, TGF-beta, and SMAD-2/3 Signaling Regulates Interferon-beta Response in Respiratory Syncytial Virus Infected Macrophages. Front. Cell. Infect. Microbiol. 2016, 6, 174. [Google Scholar] [CrossRef]
- Houghton, C.A.; Fassett, R.G.; Coombes, J.S. Sulforaphane and Other Nutrigenomic Nrf2 Activators: Can the Clinician’s Expectation Be Matched by the Reality? Oxid. Med. Cell. Longev. 2016, 2016, 7857186. [Google Scholar] [CrossRef]
- Noah, T.L.; Zhang, H.; Zhou, H.; Glista-Baker, E.; Muller, L.; Bauer, R.N.; Meyer, M.; Murphy, P.C.; Jones, S.; Letang, B.; et al. Effect of broccoli sprouts on nasal response to live attenuated influenza virus in smokers: A randomized, double-blind study. PLoS ONE 2014, 9, e98671. [Google Scholar] [CrossRef]
- Dai, J.; Gu, L.; Su, Y.; Wang, Q.; Zhao, Y.; Chen, X.; Deng, H.; Li, W.; Wang, G.; Li, K. Inhibition of curcumin on influenza A virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-kappaB pathways. Int. Immunopharmacol. 2018, 54, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Chen, D.Y.; Wen, H.W.; Ou, J.L.; Chiou, S.S.; Chen, J.M.; Wong, M.L.; Hsu, W.L. Inhibition of enveloped viruses infectivity by curcumin. PLoS ONE 2013, 8, e62482. [Google Scholar] [CrossRef] [PubMed]
- Yageta, Y.; Ishii, Y.; Morishima, Y.; Ano, S.; Ohtsuka, S.; Matsuyama, M.; Takeuchi, K.; Itoh, K.; Yamamoto, M.; Hizawa, N. Carbocisteine reduces virus-induced pulmonary inflammation in mice exposed to cigarette smoke. Am. J. Respir. Cell Mol. Biol. 2014, 50, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, A.A.; Bullen, C.K.; Villabona-Rueda, A.F.; Thompson, E.A.; Turner, M.L.; Merino, V.F.; Yan, Y.; Kim, J.; Davis, S.L.; Komm, O.; et al. Sulforaphane exhibits antiviral activity against pandemic SARS-CoV-2 and seasonal HCoV-OC43 coronaviruses in vitro and in mice. Commun. Biol. 2022, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Brandtoft, A.M.; Gunderstofte, C.; Villadsen, N.L.; Krapp, C.; Thielke, A.L.; Laustsen, A.; Peri, S.; Hansen, A.L.; Bonefeld, L.; et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 2018, 9, 3506. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008, 58, 262–270. [Google Scholar] [CrossRef]
- Mangla, B.; Javed, S.; Sultan, M.H.; Kumar, P.; Kohli, K.; Najmi, A.; Alhazmi, H.A.; Al Bratty, M.; Ahsan, W. Sulforaphane: A review of its therapeutic potentials, advances in its nanodelivery, recent patents, and clinical trials. Phytother. Res. 2021, 35, 5440–5458. [Google Scholar] [CrossRef]
- Liu, J.; Huang, C.; Liu, J.; Meng, C.; Gu, Q.; Du, X.; Yan, M.; Yu, Y.; Liu, F.; Xia, C. Nrf2 and its dependent autophagy activation cooperatively counteract ferroptosis to alleviate acute liver injury. Pharmacol. Res. 2023, 187, 106563. [Google Scholar] [CrossRef]
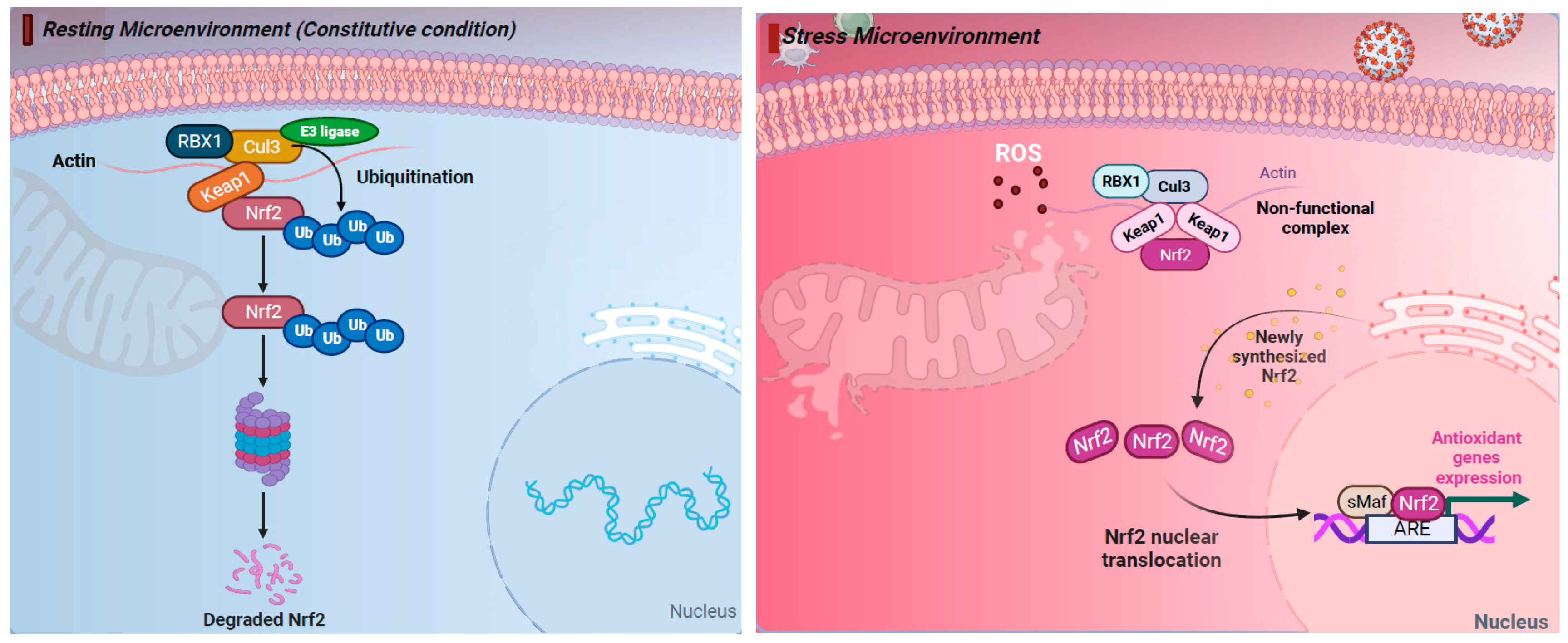
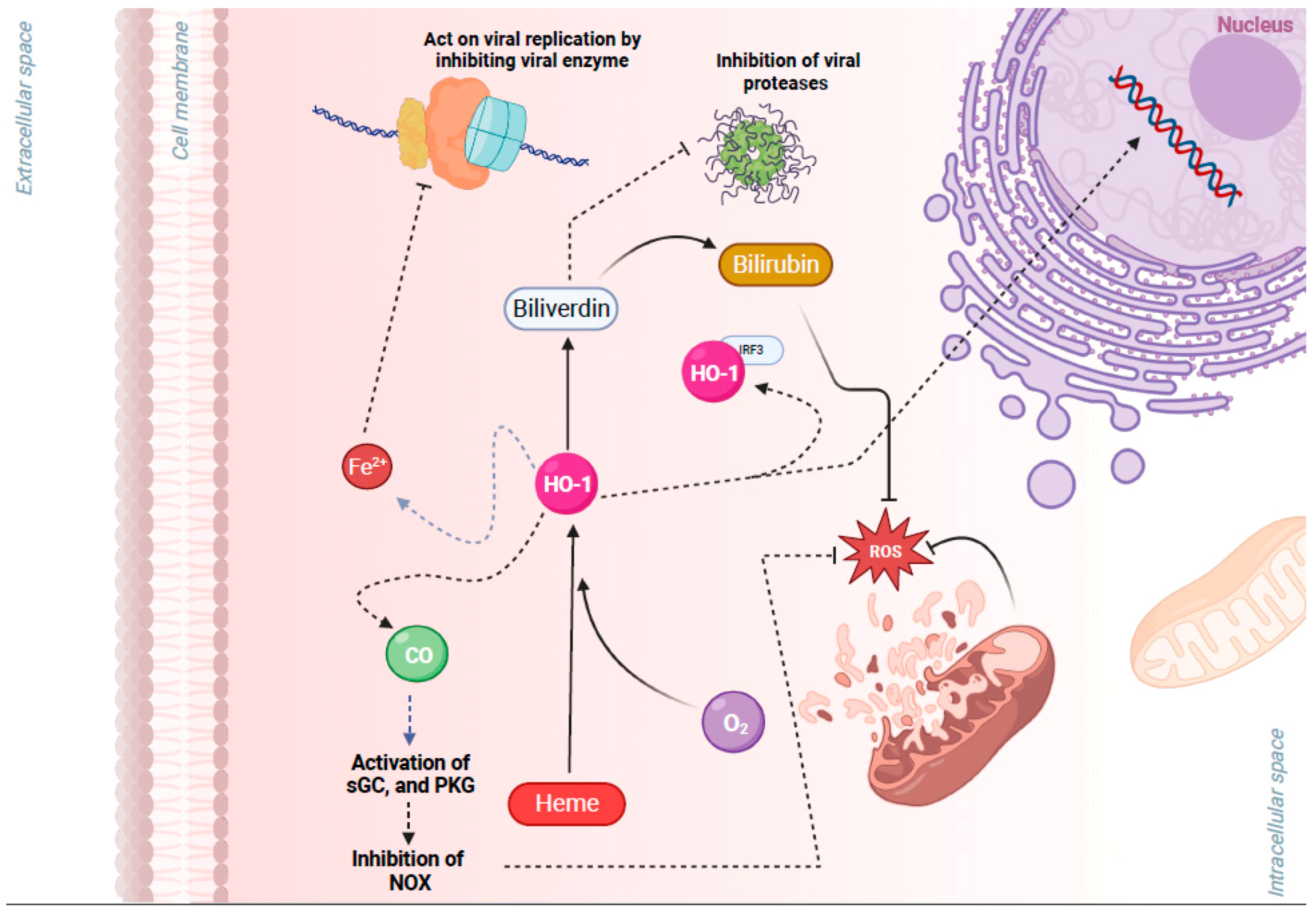

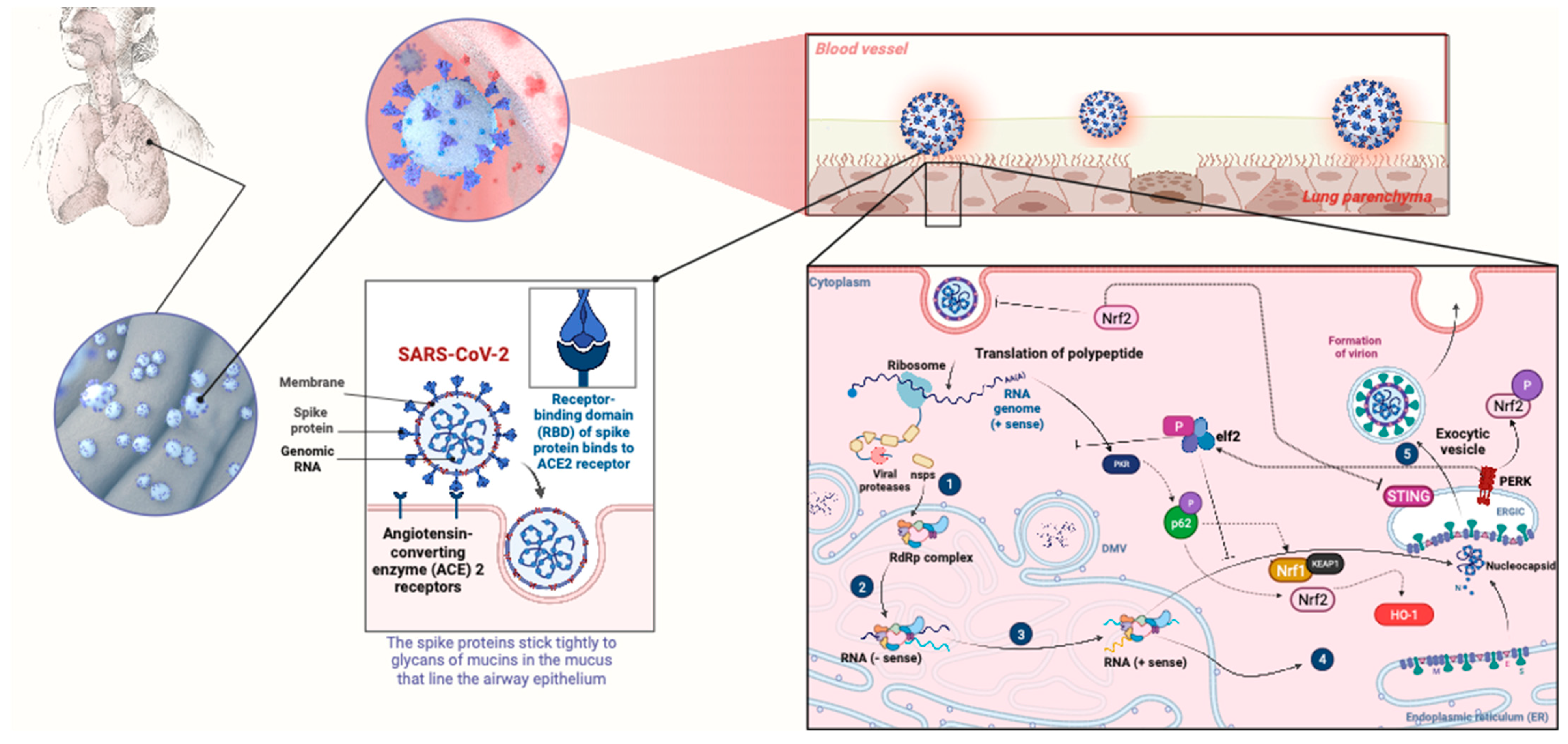
| Viruses | Mechanism of NRF2 Activation | Reference |
|---|---|---|
| Several (respiratory) viruses | ∙ ↑ ROS and mito-ROS ↑ ARE elements and phosphorylation of p62. | [5,8,9] |
| Influenza | ∙ IV strains are thought to activate the NRF2/ARE defense pathway in vitro and in mice by inducing oxidative stress and nuclear translocation and transcriptional activity of NRF2 because transcription of the NRF2 target gene HO-1 was shown to be augmented. | [12,94] |
| SARS-CoV-2 | ∙ SARS-CoV-2 infection ↓ levels of NRF2 in epithelial cells in vitro [19,20,22]. ∙ NRF2 was ↓ in RNA Seq analysis of lung biopsies of COVID-19 patients [22]. ∙ NRF2 deficiency ↑ ACE2, enhancing viral entry and as a result viral replication [19,20]. ∙ The nonstructural SARS-CoV-2 NSP14 viral protein inhibits NRF2 through ↓ of SIRT1 [95]. ∙ The SARS-CoV-2 ORF3a viral protein recruits KEAP-1 which inhibits NRF2 activity, thereby facilitating ferroptosis through the built-up ROS and the downregulation of genes like HO-1 and NQO1 [96]. ∙ The SARS-CoV-2 ORF3a viral protein binds to human HO-1 protein in vitro [24]. | [19,20,22,24,95,96] |
| RSV | ∙ RSV deregulates the NRF2 expression and its activity along with the upregulation of downstream ARE-responsive genes [98]. ∙ RSV ↓ mRNA levels of NRF2 in airway epithelial cells [27]. ∙ RSV ↑ NRF2 deacetylation, ubiquitination, and degradation through a proteasome-dependent pathway in a SUMO-specific E3 ubiquitin ligase RNF4-dependent manner. ∙ Another possible mechanism of RSV-associated NRF2 activation is the activation of [31], which activates the NRF2 pathway through direct alkylation of the NRF2 partner—KEAP1 [32]. | [27,31,32,98] |
| Rhinovirus | ∙ Rhinovirus RNA stimulates the innate immune sensor RIG-I within airway epithelial cells and activates the antiviral interferon response (greater activation in nasal cells than in bronchial cells) and the NRF2-mediated response to oxidative stress (greater activation in bronchial cells compared to nasal cells) [43]. ∙ However, the inhibitory effects were reversed in cells pretreated with the antioxidant, N-acetyl cysteine. Moreover, the secretion of anti-viral interferons ↑ in cells treated with the NRF2 agonist sulforaphane but ↓ in cells where NRF2 was silenced [44]. | [43,44] |
| Enterovirus 71 (EV71) | ∙ EV71 ↑ KEAP1 and ↓ NRF2 [47]. | [47] |
| RSV, influenza, coronaviruses, HCV | ∙ ↑ phosphorylation of the redox-sensitive PKC ↑ NRF2 dissociation from KEAP1. | [10,18,33] |
| Virus | Role of NRF2 in Viral Replication | Reference |
|---|---|---|
| Influenza virus | ∙ Activation of NRF2 leads to ↓ viral replication through interferon host responses ∙ Downregulation of NRF2 results in major ↑ of viral entry and, subsequently, viral replication. ∙ Inhibition of viral replication, growth, and protein expression after activation of NRF2. Influenza also regulates autophagy, which interacts with NRF2 and is involved in influenza replication [99]. | [11,12,13] |
| Coronaviruses | ∙ NRF2 deficiency upregulates ACE2, ↑ viral entry, and, as a result, viral replication. ∙ NRF2 induced production of HO-1 and generated Fe+2, which binds to the RNA- dependent RNA polymerase of SARS-CoV-2, inhibiting its activity and thus viral replication. ∙ NRF2 agonists like 4-OI and DMF inhibit SARS-CoV-2 replication. ∙ Vero cells infected with SARS-CoV-2 and transfected with siRNA to silence KEAP-1, thereby activating NRF2, had a decreased viral load. ∙ Absence of NRF2 in knockout mice ↑ the severity of SARS-CoV-2 infection and viral replication. | [19,20,21,22,23] |
| RSV | ∙ NRF2 knockout mice showed significantly ↑ viral titers in the lungs. ∙ Treatment of the NRF2 agonist sulforaphane on NRF2−/− and NRF2+/+ mice before RSV infection ↓ virus replication, but this significant effect was not observed in NRF2−/− mice [37]. ∙ Compared to wild-type mice, RSV-infected NRF2 KO had ↓ antioxidant enzymes and enzymes in the airway, which modulated the endogenous hydrogen sulfide (H2S) pathway that has a significant antiviral function [34]. ∙ Inducers of the NRF2-ARE pathway, such as BHA treatment, ↑ the viral clearance in murine lungs [35]. | [34,35,37] |
| Rhinovirus | ∙ Silence of NRF2 in cells led to a ↓ in the secretion of antiviral interferons and higher viral titers. | [44] |
| Enterovirus 71 (EV71) | ∙ Silencing of NRF2 is beneficial for viral replication. ∙ Activating NRF2 through downregulation of KEAP1 led to ↓ viral replication in RD cells. | [47] |
| Metapneumovirus | ∙ ↓ of NRF2-dependent genes ↑ viral replication and clinical disease upon hMPV infection. | [34] |
| Parainfluenza viruses | ∙ Cotreatment or post infection treatment with curcumin, ↓ the expression of HN viral protein, indicating that curcumin may ↓ viral entry affecting viral replication and subsequently different steps in viral replication ∙ Curcumin, an NRF2 activator, led to ↓ of F-actin, ↓ the formation of viral IBs, and ↓ viral replication. ∙ Curcumin ↓ HPIV3 replication by ↓ the endogenous PI4KB level in the cells, and ↓ the colocalization of PI4KB and IBs, affecting IB formation. | [51,52] |
| Viruses/NRF2 | HO-1 Antiviral Activity | Reference |
|---|---|---|
| Influenza | ∙ ↑ expression of HO-1 leads to ↓ viral replication during infection from Influenza A, through the upregulation of IFN- α/β and ISGs. | [14] |
| RSV | ∙ Harmacological activation of HO-1 by CoPP ↓ viral replication of RSV in lung cells of infected mice via induction of IFN-α/β expression. ∙ In vitro data suggest that ↑ of HO-1 can moderate the susceptibility of cells to hRSV infection [36]. | [36] |
| HCV | ∙ Type 1 IFN-dependent anti-HCV activity due to ↑ levels of HO-1 resulting from the usage of HO-1 agonists/inducers. ∙ Iron stopped viral replication of HCV by direct binding to the Mg+2 binding pocket of the RNA polymerase of the virus. | [88,89,90,91,92] |
| Coronaviruses | ∙ Fe+2 binds to the RNA-dependent RNA polymerase of SARS-CoV-2 inhibiting its activity and ↓ viral replication. ORF3a protein binds to human HMOX1 protein in vitro [24]. | [19] |
| EV71 | ∙ The overexpression of HO-1 ↓ NADPH oxidase/ROS production that is induced by enterovirus 71 and hence ↓ viral replication. This effect was abolished if cells were pretreated with zinc–protoporphyrin IX, an HO-1 activity inhibitor. ∙ Bilirubin has also been found to exert antiviral activity against EV71 reducing its replication and as a result infectivity in vitro. | [48,49] |
| HIV | ∙ BV and BR have been identified to act as inhibitors for the protease of HIV, interfering with the life cycle of the virus. | [93] |
| Virus | Role of NRF2 in Inflammation | References |
|---|---|---|
| Influenza virus | ∙ Inactivation of NF-κΒ transcription factor. ∙ ↓ of NF-κΒ-mediated inflammation and associated lung permeability damage, mucus hypersecretion, lung permeability damage, as well as mucus hypersecretion, through reduced NF-κΒ-mediated inflammation and associated proinflammatory cytokines [15]. ∙ Induces anti-inflammatory effects in vivo through the HO-1 pathway [100,101]. ∙ NLRP3 activation form PB1-F2 influenza A protein. ∙ K+ efflux and ROS dependent activation of NLRP3 inflammasome ∙ Impacts function of alveolar macrophages (AMϕ) that are important essential for preventing respiratory failure and mortality after infection from influenza virus in mice [17]. ∙ Attenuates virus-induced inflammation through increased GSH levels and IL-8 secretion in ATI-like cells (alveolar epithelial cells) in vitro [12]. | [15,16,17] |
| Coronavirus | ∙ NRF2 is directly able to inhibit IL6, IL-1B, a key hallmark of the cytokine storm in SARS-CoV-2 infection. ∙ Absence of NRF2 in knockout mice ↑ the severity of SARS-CoV-2 infection, pulmonary inflammation. ∙ In humans, SNPs in the Nrf2 gene promoter region can determine susceptibility to respiratory failure with COPD, indicating the importance of NRF2 in pulmonary inflammation. ∙ Cytokine storm due to T cell depletion and widespread pulmonary inflammation. ∙ Contradictory effect on proinflammatory nature of factors like NF-kB. | [23,25,26] |
| RSV | ∙ Severe inflammation in NRF2−/− mice compared to NRF2+/+ mice. ∙ RSV-infected NRF2 KO mice are reported to have a significant ↑ in airway neutrophilia and inflammatory cytokines. ∙ ↓ lung inflammation when pretreated with sulforaphane. ∙ ↓ ROS- and K+ efflux-dependent activation of NLRP3 inflammasome. ∙ SH viroporin activates NLRP3 inflammasome. ∙ Impacts function of alveolar macrophages (AMϕ), which are important to attenuate virus-induced inflammation. | [37,38,39,40,102] |
| Metapneumovirus | ∙ NRF2 KO mice infected with hMPV had ↓ expression of antioxidant enzymes (AOE) and ↑ viral-mediated oxidative stress and airway damage compared to NRF2+/+ mice. | [34] |
| Enterovirus 71 | ∙ By silencing KEAP1, the induced ROS, apoptosis, and inflammation was ↓ in the EV71 infected cells. However, when both KEAP1 and NRF2 were silenced in Vero and RD cells, these effects were restored. ∙ Inflammation-promoting cytokines and chemokines influence the severity of the EV71 infection. | [47,50] |
| Rhinovirus | ∙ 2B viroporin activates NLRP3 inflammasome | [16,45] |
| Viruses/NRF2 | Impact on Apoptosis | Reference |
|---|---|---|
| Adenoviruses | ∙ Complex effects. ∙ ↑ apoptosis: ↑ sensitivity to TNFa that induces apoptosis, ↑ PP2A, and ↑ p53. ∙ ↓ apoptosis through several mechanisms: interacts with FADD, ↓ CD95-mediated apoptosis, ↓ phospholipase A2, ↓ Fas, ↓ p53, and ↓ pro-apoptotic proteins of the Bcl-2 family, such as Bax, Bak, BNIP3, and Bnip3L. ∙ ↓ apoptosis of the host cell in order to ↑ efficiently and the capacity of the virus to ‘hijack’ host cell apoptotic machinery. | [103,104,105,106] |
| RSV | ∙ ↑ interferons and caspase 1. ∙ Experimental studies have shown that autophagy plays a very crucial role in RSV replication [107]. | [105] |
| Influenza | ∙ ↑ Fas expression. ∙ ↓ PKR and apoptosis. ∙ Apoptosis plays a role in viral release. | [105,106] |
| Rhinovirus, enteroviruses | ∙ ↑ apoptosis through unknown mechanism. | [105] |
| Coronaviruses | ∙ ↑ apoptosis through ORF proteins and unknown mechanisms. | [105] |
| Viruses/NRF2 | Impact on Ferroptosis | Reference |
|---|---|---|
| NRF2 | ∙ NRF2 ↓ ROS and ↑ antioxidant responses, and ↑ GPX4-induced ↓ of ferroptosis. ∙ NRF2 ↑ Heme Oxygenase 1 (HO-1) that ↓ ferroptosis. ∙ NRF2 ↑ antioxidant enzymes. | [9] |
| Influenza | ∙ Iron ↓ viral genome amplification and viral replication. ∙ Influenza ↓ cellular GSH and/or affects GPX4 activity. ∙ Neuraminidase of Influenza A binds lysosome-associated membrane proteins and ↑ lysosome rupture. | [9,108,109,110,111,112] |
| SARS-CoV-2, SARS-CoV, other coronaviruses | ∙ SARS-CoV-2 Potentially causes cellular iron overload and iron scavenging. ∙ SARS-CoV-2 ↑serum ferritin concentration. ∙ CoVs↓ cellular GSH and/or affect GPX4 activity. ∙ SARS-CoV ORF-3a viral protein ↑ lysosomal damage and dysfunction. | [113,114,115,116,117] |
| RSV | ∙ ↑ the expression of 12/15-LOX and mitochondrial iron content. | [118] |
| EV-71, CB3 | ∙ Iron ↓ viral genome amplification and viral replication of EV-71. ∙ CB3 ↑the expression NRAMP (DMT) and ↑cellular iron uptake. | [108,119,120] |
| Non-respiratory viruses: HBV, HCV, WNV, Dengue virus, HSV, KSHV | ∙ HBV, HCV: ↑ serum and cellular iron uptake and ↓ hepcidin expression, ↑ serum ferritin concentration, and uses TfR1 as a cellular receptor. ∙ HIV ↓ serum iron, ↑ the expression of hepcidin, ↑cellular iron via hepcidin mediated degradation of ferroportin, ↓ cellular GSH and/or affects GPX4 activity, and upregulates the expression of system xc-. ∙ WNV ↑ the expression NRAMP (DMT) and ↑cellular iron uptake. ∙ Dengue virus ↓ cellular GSH and/or affects GPX4 activity. ∙ HSV ↓ cellular GSH and/or affects GPX4 activity. ∙ JEV ↓ cellular GSH and/or affects GPX4 activity, produces lipid peroxide free radicals, and ↑ the expression of system xc-. ∙ KSHV ↓ cellular GSH and/or affects GPX4 activity. ∙ Zika virus ↓ cellular GSH and/or affects GPX4 activity. ∙ Other viruses (e.g., hemorrhagic viruses) use NRAMP or TfR1 as a cellular receptor. | [121] |
| NRF2 Activator | Impact | Reference |
|---|---|---|
| 4-OI | ∙ ↓ viral replication of SARS-CoV-2. | [188] |
| DMF | ∙ Inhibition of SARS-CoV-2 viral replication. | [188] |
| SFN | ∙ Anti-SARS-CoV-2 properties through induction of NQO1 activation. ∙ Anti-viral properties against RSV. ∙ ↓ in viral load and IL-6 in influenza infection. | [37,189,190,194] |
| Curcumin | ∙ Inhibition of influenza, parainfluenza, and RSV viral replication. ∙ ↓ in oxidative stress through HO-1 activation. | [51,191,192] |
| EGCG | ∙ Anti-influenza activity through blockage of viral entry and subsequently viral replication. | [11] |
| Carbocistein | ∙ ↓ in the expression of the nucleoprotein of influenza virus and thus viral replication. | [193] |
| BHA | ∙ Severe ↓ in RSV-induced oxidative stress. | [168] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daskou, M.; Fotooh Abadi, L.; Gain, C.; Wong, M.; Sharma, E.; Kombe Kombe, A.J.; Nanduri, R.; Kelesidis, T. The Role of the NRF2 Pathway in the Pathogenesis of Viral Respiratory Infections. Pathogens 2024, 13, 39. https://doi.org/10.3390/pathogens13010039
Daskou M, Fotooh Abadi L, Gain C, Wong M, Sharma E, Kombe Kombe AJ, Nanduri R, Kelesidis T. The Role of the NRF2 Pathway in the Pathogenesis of Viral Respiratory Infections. Pathogens. 2024; 13(1):39. https://doi.org/10.3390/pathogens13010039
Chicago/Turabian StyleDaskou, Maria, Leila Fotooh Abadi, Chandrima Gain, Michael Wong, Eashan Sharma, Arnaud John Kombe Kombe, Ravikanth Nanduri, and Theodoros Kelesidis. 2024. "The Role of the NRF2 Pathway in the Pathogenesis of Viral Respiratory Infections" Pathogens 13, no. 1: 39. https://doi.org/10.3390/pathogens13010039
APA StyleDaskou, M., Fotooh Abadi, L., Gain, C., Wong, M., Sharma, E., Kombe Kombe, A. J., Nanduri, R., & Kelesidis, T. (2024). The Role of the NRF2 Pathway in the Pathogenesis of Viral Respiratory Infections. Pathogens, 13(1), 39. https://doi.org/10.3390/pathogens13010039











