An In Silico Functional Analysis of Non-Synonymous Single-Nucleotide Polymorphisms of Bovine CMAH Gene and Potential Implication in Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of nsSNPs in bCMAH from the 1000 Bull Genomes Sequence Data
2.2. Prediction, Refinement, and Validation of Tertiary Structure of Bovine CMAH Protein
2.3. Evaluation of the Functional Impacts of the nsSNPs on the Function and Stability of the Bovine CMAH Protein
2.4. Sequence Conservational Analysis
2.5. Post-Translational Modification Sites’ Prediction
2.6. Stability Analysis
2.7. Active Site Prediction and Molecular Dynamic Simulation
3. Results
3.1. Distribution of nsSNPs in Different Breeds Analysed in 1000 Bull Genomes Sequence Data
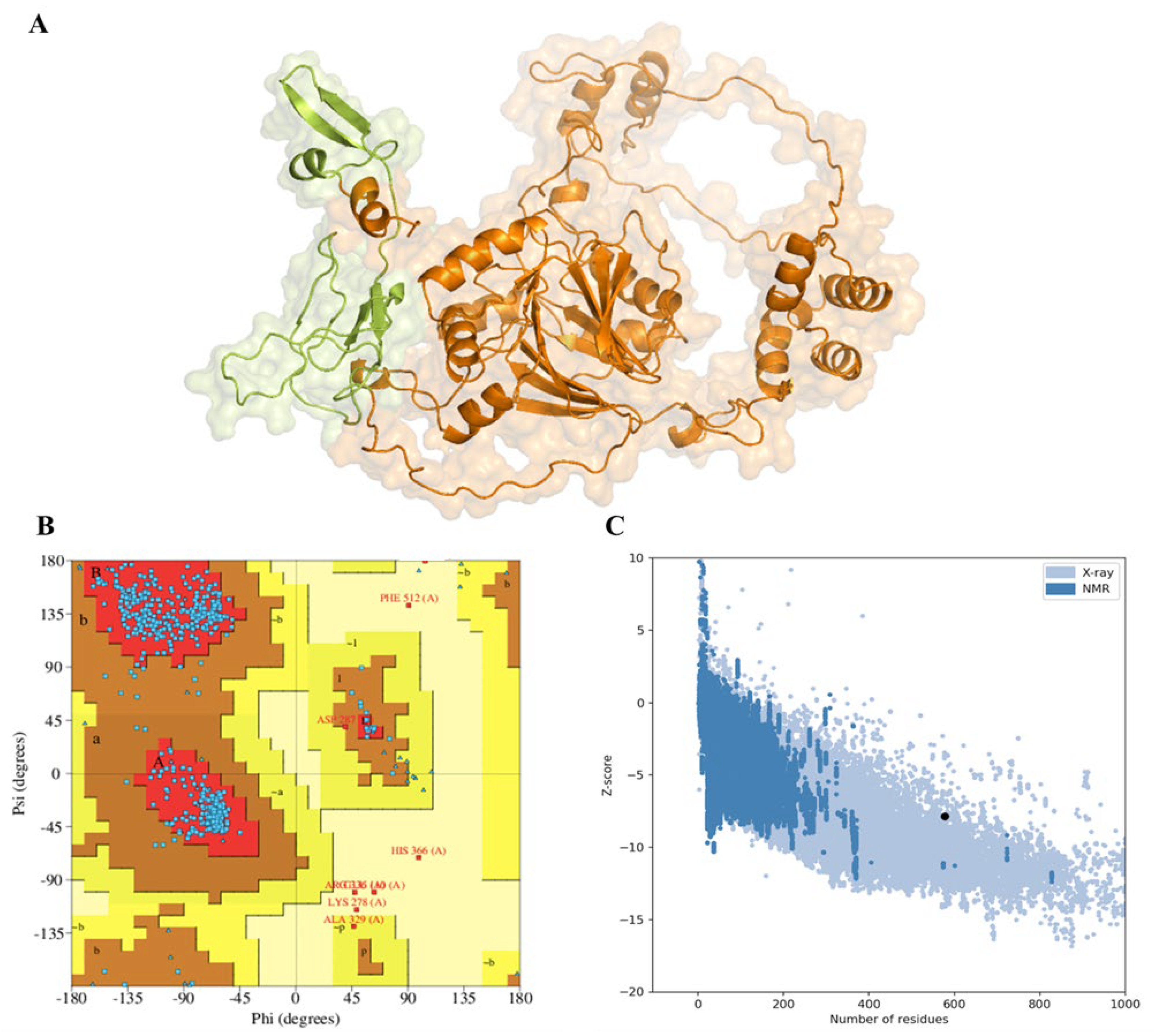
3.2. Secondary and Tertiary Structure Prediction and Validation
3.3. Prediction of Pathogenic and Damaging Amino Acids of Bovine CMAH Protein
3.4. Prediction of the Effects of Amino Acid Substitutions on Bovine CMAH Protein Stability
3.5. Sequence Conservational Analysis and the Predicted PTMs
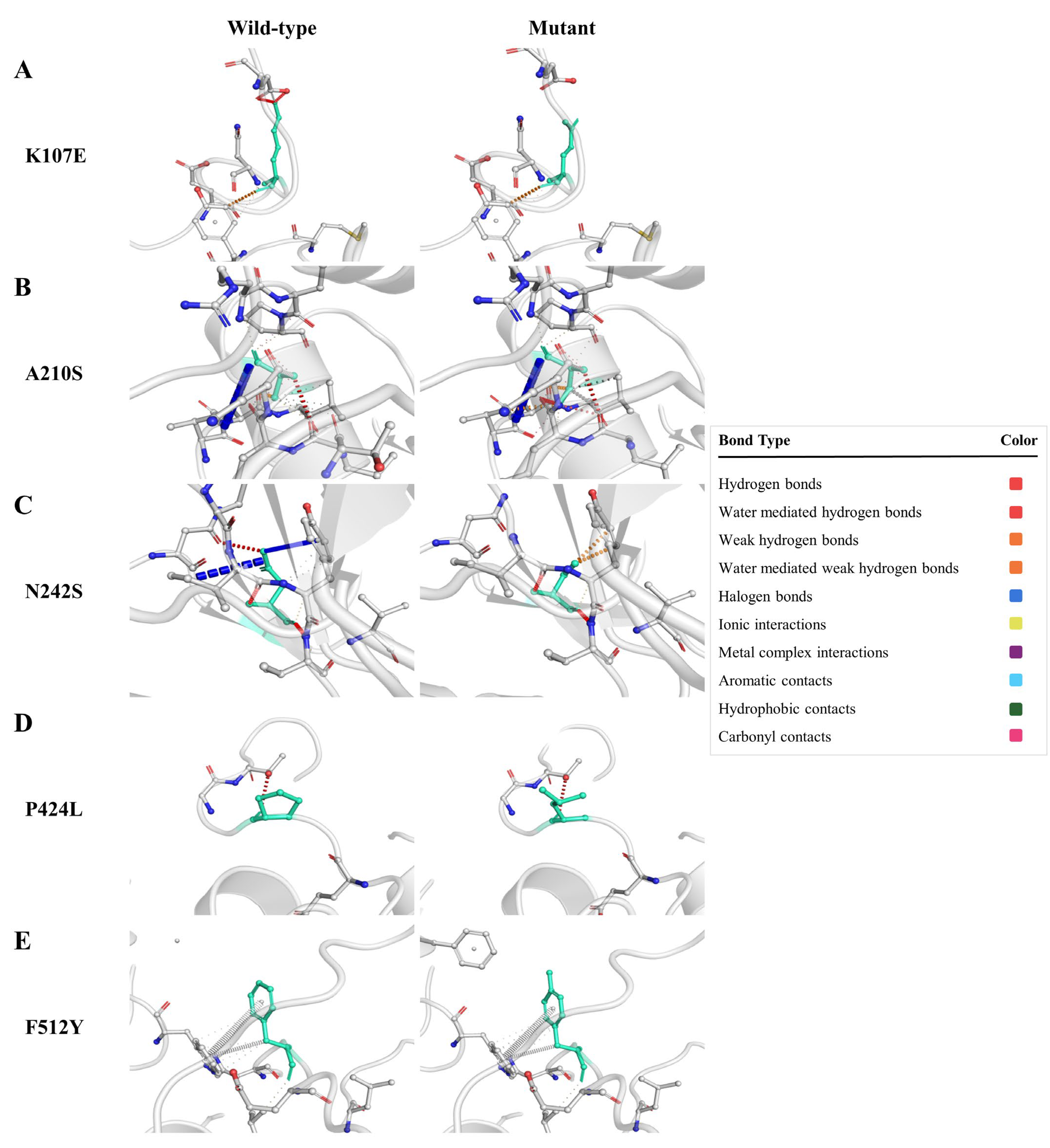
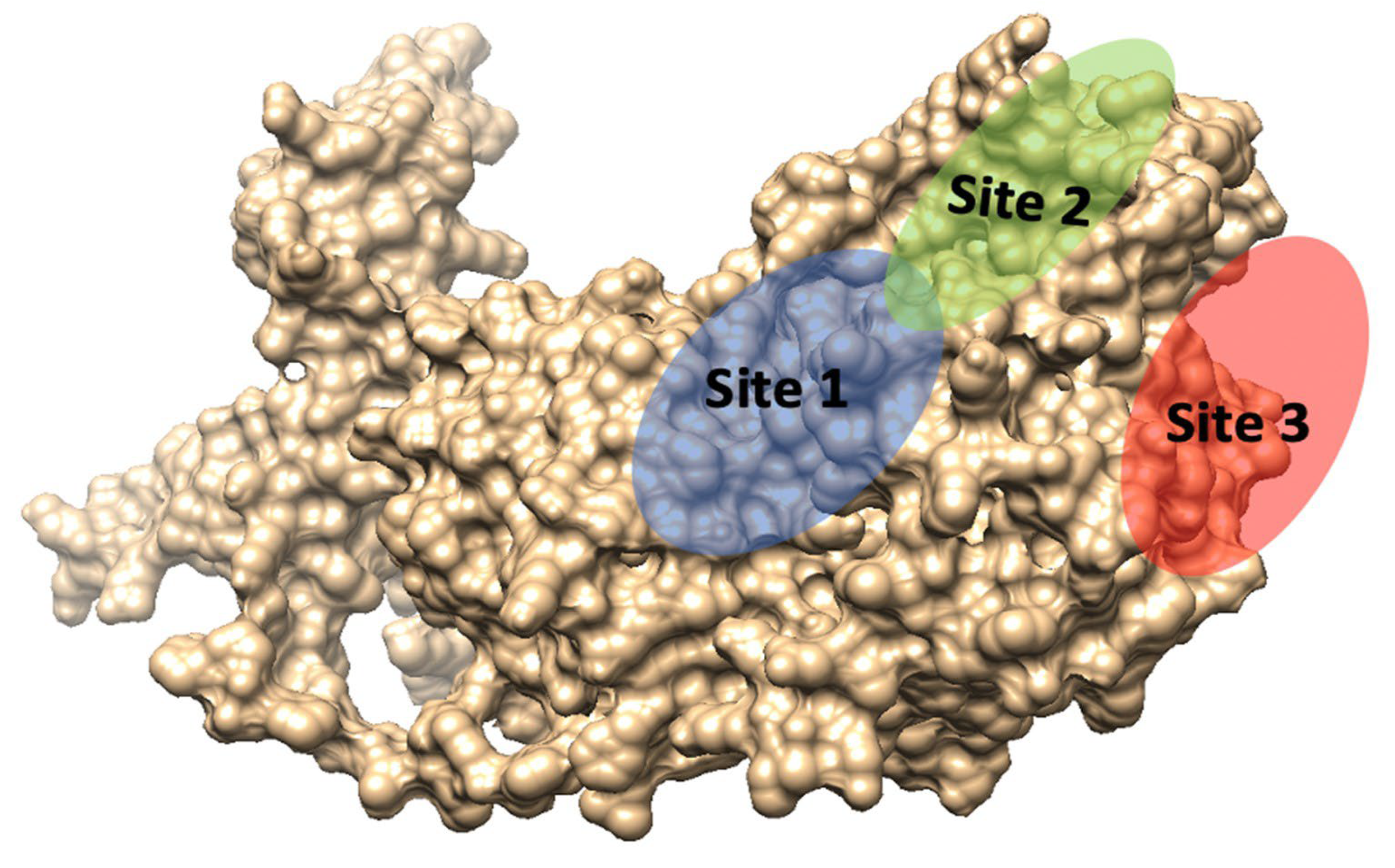
3.6. Active Sites’ Prediction and Molecular Dynamics Simulations
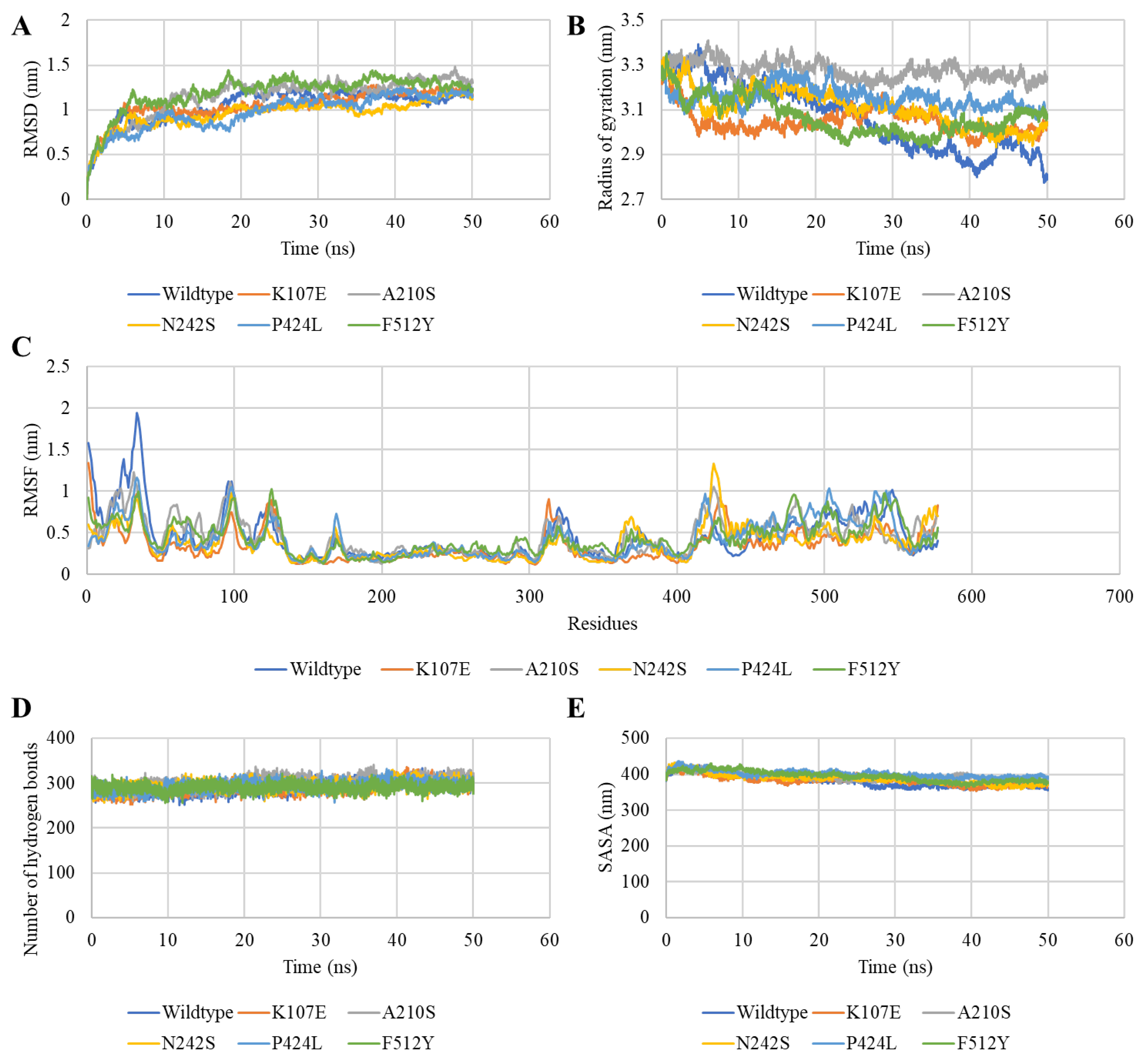
3.6.1. Bovine CMAH Mutations’ Impact on Its Structural Stability
3.6.2. Mutations Elicited Structural Distortion in Bovine CMAH Protein
4. Discussion
5. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varki, A. Uniquely human evolution of sialic acid genetics and biology. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S2), 8939–8946. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; Varki, A. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem. Rev. 2002, 102, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Kooner, A.S.; Yu, H.; Chen, X. Synthesis of N-glycolylneuraminic acid (Neu5Gc) and its glycosides. Front. Immunol. 2019, 10, 2004. [Google Scholar] [CrossRef] [PubMed]
- Dhar, C.; Sasmal, A.; Varki, A. From “serum sickness” to “xenosialitis”: Past, present, and future significance of the non-human sialic acid Neu5Gc. Front. Immunol. 2019, 10, 807. [Google Scholar] [CrossRef]
- Magre, S.; Takeuchi, Y.; Bartosch, B. Xenotransplantation and pig endogenous retroviruses. Rev. Med. Virol. 2003, 13, 311–329. [Google Scholar] [CrossRef]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic acid receptors of viruses. In Sialoglyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans; Springer: Cham, Switzerland, 2015; pp. 1–28. [Google Scholar]
- Payne, S. Viruses: From Understanding to Investigation; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Varki, A.; Schauer, R. Sialic acids. In Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring, NY, USA, 2009. [Google Scholar]
- Delorme, C.; Brüssow, H.; Sidoti, J.; Roche, N.; Karlsson, K.-A.; Neeser, J.-R.; Teneberg, S. Glycosphingolipid binding specificities of rotavirus: Identification of a sialic acid-binding epitope. J. Virol. 2001, 75, 2276–2287. [Google Scholar] [CrossRef]
- Kyogashima, M.; Ginsburg, V.; Krivan, H.C. Escherichia coli K99 binds to N-glycolylsialoparagloboside and N-glycolyl-GM3 found in piglet small intestine. Arch. Biochem. Biophys. 1989, 270, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Schwegmann, C.; Zimmer, G.; Yoshino, T.; Enss, M.-L.; Herrler, G. Comparison of the sialic acid binding activity of transmissible gastroenteritis coronavirus and E. coli K99. Virus Res. 2001, 75, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Abe, K.; Nakazawa, M.; Naiki, M. Ganglioside epitope recognized by K99 fimbriae from enterotoxigenic Escherichia coli. Infect. Immun. 1989, 57, 907–911. [Google Scholar] [CrossRef]
- Teneberg, S.; Willemsen, P.; de Graaf, F.K.; Karlsson, K.-A. Receptor-active glycolipids of epithelial cells of the small intestine of young and adult pigs in relation to susceptibility to infection with Escherichia coli K99. FEBS Lett. 1990, 263, 10–14. [Google Scholar] [CrossRef]
- Wasik, B.R.; Barnard, K.N.; Parrish, C.R. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol. 2016, 24, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Dang, V.T.; Fleming, F.E.; von Itzstein, M.; Coulson, B.S.; Blanchard, H. Structural basis of rotavirus strain preference toward N-acetyl-or N-glycolylneuraminic acid-containing receptors. J. Virol. 2012, 86, 13456–13466. [Google Scholar] [CrossRef] [PubMed]
- Consortium, B.H.; Gibbs, R.A.; Taylor, J.F.; Van Tassell, C.P.; Barendse, W.; Eversole, K.A.; Gill, C.A.; Green, R.D.; Hamernik, D.L.; Kappes, S.M. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 2009, 324, 528–532. [Google Scholar] [CrossRef]
- Ghosh, M.; Sharma, N.; Singh, A.K.; Gera, M.; Pulicherla, K.K.; Jeong, D.K. Transformation of animal genomics by next-generation sequencing technologies: A decade of challenges and their impact on genetic architecture. Crit. Rev. Biotechnol. 2018, 38, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Coppieters, W.; Rollin, F.; Desmecht, D.; Agerholm, J.S.; Cambisano, N.; Carta, E.; Dardano, S.; Dive, M.; Fasquelle, C. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat. Genet. 2008, 40, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Schaub, M.A.; Boyle, A.P.; Kundaje, A.; Batzoglou, S.; Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012, 22, 1748–1759. [Google Scholar] [CrossRef]
- Van der Spek, D.; Van Arendonk, J.; Bovenhuis, H. Genome-wide association study for claw disorders and trimming status in dairy cattle. J. Dairy Sci. 2015, 98, 1286–1295. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genom. Hum. Genet. 2006, 7, 61–80. [Google Scholar] [CrossRef]
- Rafaee, A.; Kashani-Amin, E.; Meybodi, A.M.; Ebrahim-Habibi, A.; Sabbaghian, M. Structural modeling of human AKAP3 protein and in silico analysis of single nucleotide polymorphisms associated with sperm motility. Sci. Rep. 2022, 12, 3656. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, C.; Wang, Z.; Lv, H.; Li, X. In silico analysis of non-synonymous single nucleotide polymorphisms (nsSNPs) in the human GJA3 gene associated with congenital cataract. BMC Mol. Cell Biol. 2020, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Bighignoli, B.; Niini, T.; Grahn, R.A.; Pedersen, N.C.; Millon, L.V.; Polli, M.; Longeri, M.; Lyons, L.A. Cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) mutations associated with the domestic cat AB blood group. BMC Genet. 2007, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Grahn, R.A.; Gustafson, N.A.; Proverbio, D.; Spada, E.; Adhikari, B.; Cheng, J.; Andrews, G.; Lyons, L.A.; Helps, C.R. A novel variant in CMAH is associated with blood type AB in Ragdoll cats. PLoS ONE 2016, 11, e0154973. [Google Scholar] [CrossRef]
- Uno, Y.; Kawakami, S.; Ochiai, K.; Omi, T. Molecular characterization of cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) associated with the erythrocyte antigens in dogs. Canine Genet. Epidemiol. 2019, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Spruit, C.M.; Nemanichvili, N.; Okamatsu, M.; Takematsu, H.; Boons, G.-J.; de Vries, R.P. N-glycolylneuraminic acid in animal models for human influenza A virus. Viruses 2021, 13, 815. [Google Scholar] [CrossRef] [PubMed]
- Alisson-Silva, F.; Liu, J.Z.; Diaz, S.L.; Deng, L.; Gareau, M.G.; Marchelletta, R.; Chen, X.; Nizet, V.; Varki, N.; Barrett, K.E. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathol. 2018, 14, e1007133. [Google Scholar] [CrossRef]
- Reuven, E.M.; Leviatan Ben-Arye, S.; Marshanski, T.; Breimer, M.E.; Yu, H.; Fellah-Hebia, I.; Roussel, J.C.; Costa, C.; Galiñanes, M.; Mañez, R. Characterization of immunogenic Neu5Gc in bioprosthetic heart valves. Xenotransplantation 2016, 23, 381–392. [Google Scholar] [CrossRef]
- TruSeq DNA PCR-Free | Simple Prep for Sequencing Complex Genomes. Available online: https://www.illumina.com/products/by-type/sequencing-kits/library-prep-kits/truseq-dna-pcr-free.html (accessed on 5 April 2023).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatic 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Hayes, B.J.; Daetwyler, H.D. 1000 bull genomes project to map simple and complex genetic traits in cattle: Applications and outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Vernon, R.; Thompson, J.; Tyka, M.; Sadreyev, R.; Pei, J.; Kim, D.; Kellogg, E.; DiMaio, F.; Lange, O. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 2009, 77, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Sigrist, C.J.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012, 41, D344–D347. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2012, 41, D508–D516. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14, S6. [Google Scholar]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Shah, H.; Rehman, A.; Badshah, Y.; Ashraf, N.M.; Shabbir, M. Influence of PRKCE non-synonymous variants on protein dynamics and functionality. Hum. Mol. Genet. 2022, 31, 2236–2261. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Huang, B. MetaPocket: A meta approach to improve protein ligand binding site prediction. OMICS A J. Integr. Biol. 2009, 13, 325–330. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36: An improved force field for folded and intrinsically disordered proteins. Biophys. J. 2017, 112, 175a–176a. [Google Scholar] [CrossRef]
- Armenta, S.; Sánchez-Cuapio, Z.; Munguia, M.E.; Pulido, N.O.; Farrés, A.; Manoutcharian, K.; Hernandez-Santoyo, A.; Moreno-Mendieta, S.; Sánchez, S.; Rodríguez-Sanoja, R. The role of conserved non-aromatic residues in the Lactobacillus amylovorus α-amylase CBM26-starch interaction. Int. J. Biol. Macromol. 2019, 121, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Parolin Schnekenberg, R.; Perkins, E.M.; Miller, J.W.; Davies, W.I.; D’Adamo, M.C.; Pessia, M.; Fawcett, K.A.; Sims, D.; Gillard, E.; Hudspith, K. De novo point mutations in patients diagnosed with ataxic cerebral palsy. Brain 2015, 138, 1817–1832. [Google Scholar] [CrossRef]
- Smigielski, E.M.; Sirotkin, K.; Ward, M.; Sherry, S.T. dbSNP: A database of single nucleotide polymorphisms. Nucleic Acids Res. 2000, 28, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Teng, S.; Wang, L.; Schwartz, C.E.; Alexov, E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum. Mutat. 2010, 31, 1043–1049. [Google Scholar] [CrossRef]
- Kehl, A.; Heimberger, K.; Langbein-Detsch, I.; Boehmer, S.; Raj, K.; Mueller, E.; Giger, U. Molecular characterization of blood type A, B, and C (AB) in domestic cats and a CMAH genotyping scheme. PLoS ONE 2018, 13, e0204287. [Google Scholar] [CrossRef]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Functional advantages of dynamic protein disorder. FEBS Lett. 2015, 589, 2433–2440. [Google Scholar] [CrossRef]
- Babu, M.M.; Kriwacki, R.W.; Pappu, R.V. Versatility from protein disorder. Science 2012, 337, 1460–1461. [Google Scholar] [CrossRef]
- Omi, T.; Nakazawa, S.; Udagawa, C.; Tada, N.; Ochiai, K.; Chong, Y.H.; Kato, Y.; Mitsui, H.; Gin, A.; Oda, H. Molecular characterization of the cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH) gene associated with the feline AB blood group system. PLoS ONE 2016, 11, e0165000. [Google Scholar] [CrossRef]
- Li, S.; Iakoucheva, L.M.; Mooney, S.D.; Radivojac, P. Loss of post-translational modification sites in disease. In Biocomputing 2010; World Scientific: Singapore, 2010; pp. 337–347. [Google Scholar]
- Kotch, F.W.; Guzei, I.A.; Raines, R.T. Stabilization of the collagen triple helix by O-methylation of hydroxyproline residues. J. Am. Chem. Soc. 2008, 130, 2952–2953. [Google Scholar] [CrossRef]
- Lee, J.-W.; Bae, S.-H.; Jeong, J.-W.; Kim, S.-H.; Kim, K.-W. Hypoxia-inducible factor (HIF-1) α: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jacob, K.K.; Radhika, G.; Aravindakshan, T. An in silico evaluation of non-synonymous single nucleotide polymorphisms of mastitis resistance genes in cattle. Anim. Biotechnol. 2020, 31, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Rehman, M.U.; Ahmad, S.M.; Mehraj, T.; Hussain, I.; Nadeem, A.; Mir, M.U.R.; Ganie, S.A. In Silico Tools for Analysis of Single-Nucleotide Polymorphisms in the Bovine Transferrin Gene. Animals 2022, 12, 693. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Oh, J.-D.; Won, K.-H.; Song, K.-D. In silico approaches to identify the functional and structural effects of non-synonymous SNPs in selective sweeps of the Berkshire pig genome. Asian-Australas. J. Anim. Sci. 2018, 31, 1150. [Google Scholar] [CrossRef] [PubMed]


| Coding Exon |
cDNA Variant (XM_024984024.1) | Protein Variant (XP_024839792.1) | Genomic Position | RefSNP ID (dbSNP) |
|---|---|---|---|---|
| 4 | c.319A>G | K107E | BTA23:g.32,721,570 | rs208635220 |
| 6 | c.628G>T | A210S | BTA23:g.32,727,639 | rs435799892 |
| 6 | c.725A>G | N242S | BTA23:g.32,727,736 | rs109811989 |
| 11 | c.1271C>T | P424L | BTA23:g.32,743,918 | rs518400910 |
| 12 | c.1535T>A | F512Y | BTA23:g.32,745,866 | rs380571713 |
| Coding Exon |
cDNA Variant (XM_024984024.1) |
Heterozygous (n) | Homozygous Alternative Allele (n) | Reference Allele (n) | Null Data (n) | Total (n) |
|---|---|---|---|---|---|---|
| 4 | c.319A>G | 1091 | 362 | 1140 | 131 | 2724 |
| 6 | c.628G>T | 40 | 4 | 2662 | 18 | 2724 |
| 6 | c.725A>G | 234 | 27 | 2443 | 20 | 2724 |
| 11 | c.1271C>T | 15 | 0 | 2689 | 20 | 2724 |
| 12 | c.1535T>A | 77 | 14 | 2610 | 23 | 2724 |
|
Protein Variant (XP_024839792.1) | PolyPhen-2 | SNPs&GO | PROVEAN | SIFT | PANTHER |
|---|---|---|---|---|---|
| K107E | Benign | Neutral | Neutral | Tolerated | Probably Benign |
| A210S | Benign | Neutral | Neutral | Tolerated | Possibly Damaging |
| N242S | Benign | Neutral | Neutral | Tolerated | Probably Benign |
| P424L | Probably Damaging | Disease | Deleterious | Deleterious | Probably Damaging |
| F512Y | Benign | Neutral | Neutral | Tolerated | Probably Benign |
|
Protein Variant (XP_024839792.1) | Conservation | PTMs |
|---|---|---|
| K107E | Variable | Ub, Su, Ac, Me, Hy |
| A210S | Average | None |
| N242S | Average | None |
| P424L | Highly Conserved | Hy |
| F512Y | Highly Conserved | None |
| Site | Site Score | Size | Volume | DScore | Residues |
|---|---|---|---|---|---|
| 1 | 1.017 | 90 | 184.53 | 0.961 | Ser72, Cys75, Thr76, Asn79, Asp83, Val84, Ser85, Thr86, Met87, Lys88, Pro93, Gly94, Ser95, Phe96, Lys222, Met262, Asp263, Gly264, Ile265, His266, Pro267, Glu268, Asp270 |
| 2 | 1.016 | 214 | 619.11 | 1.057 | Tyr41, Lys42, Ser43, Leu46, Arg48, Lys51, Cys54, Lys55, Leu60, Thr163, Gly164, Pro165, Ala166, Phe167, Ala168, Gly170, Trp171, Trp172, Leu173, Leu174, His175, Pro177, Pro178, Trp181, Met196, His197, Ser198, Leu201, Ser202, Tyr203, Pro204, Lys208, Pro226, Val227, Trp229, Asn230, Leu231, Asn232, Gln233, Glu513, Glu514 |
| 3 | 0.962 | 71 | 154.69 | 1.001 | Pro381, Asp382, Leu384, Asn385, Val394, Thr396, Trp397, Thr398, Lys468, Asp469, Leu530, Leu531, Leu535 |
| Mutation | DDG | Stability |
|---|---|---|
| K107E | −0.17 | Increase |
| A210S | −0.44 | Increase |
| N242S | −0.49 | Increase |
| P424L | −0.32 | Increase |
| F512Y | −0.33 | Increase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogun, O.J.; Soremekun, O.S.; Thaller, G.; Becker, D. An In Silico Functional Analysis of Non-Synonymous Single-Nucleotide Polymorphisms of Bovine CMAH Gene and Potential Implication in Pathogenesis. Pathogens 2023, 12, 591. https://doi.org/10.3390/pathogens12040591
Ogun OJ, Soremekun OS, Thaller G, Becker D. An In Silico Functional Analysis of Non-Synonymous Single-Nucleotide Polymorphisms of Bovine CMAH Gene and Potential Implication in Pathogenesis. Pathogens. 2023; 12(4):591. https://doi.org/10.3390/pathogens12040591
Chicago/Turabian StyleOgun, Oluwamayowa Joshua, Opeyemi S. Soremekun, Georg Thaller, and Doreen Becker. 2023. "An In Silico Functional Analysis of Non-Synonymous Single-Nucleotide Polymorphisms of Bovine CMAH Gene and Potential Implication in Pathogenesis" Pathogens 12, no. 4: 591. https://doi.org/10.3390/pathogens12040591
APA StyleOgun, O. J., Soremekun, O. S., Thaller, G., & Becker, D. (2023). An In Silico Functional Analysis of Non-Synonymous Single-Nucleotide Polymorphisms of Bovine CMAH Gene and Potential Implication in Pathogenesis. Pathogens, 12(4), 591. https://doi.org/10.3390/pathogens12040591






