Enrichment of Acid-Associated Microbiota in the Saliva of Type 2 Diabetes Mellitus Adults: A Systematic Review
Abstract
1. Introduction
2. Materials & Methods
2.1. Study Design
2.2. Eligibility Criteria and Search Strategy
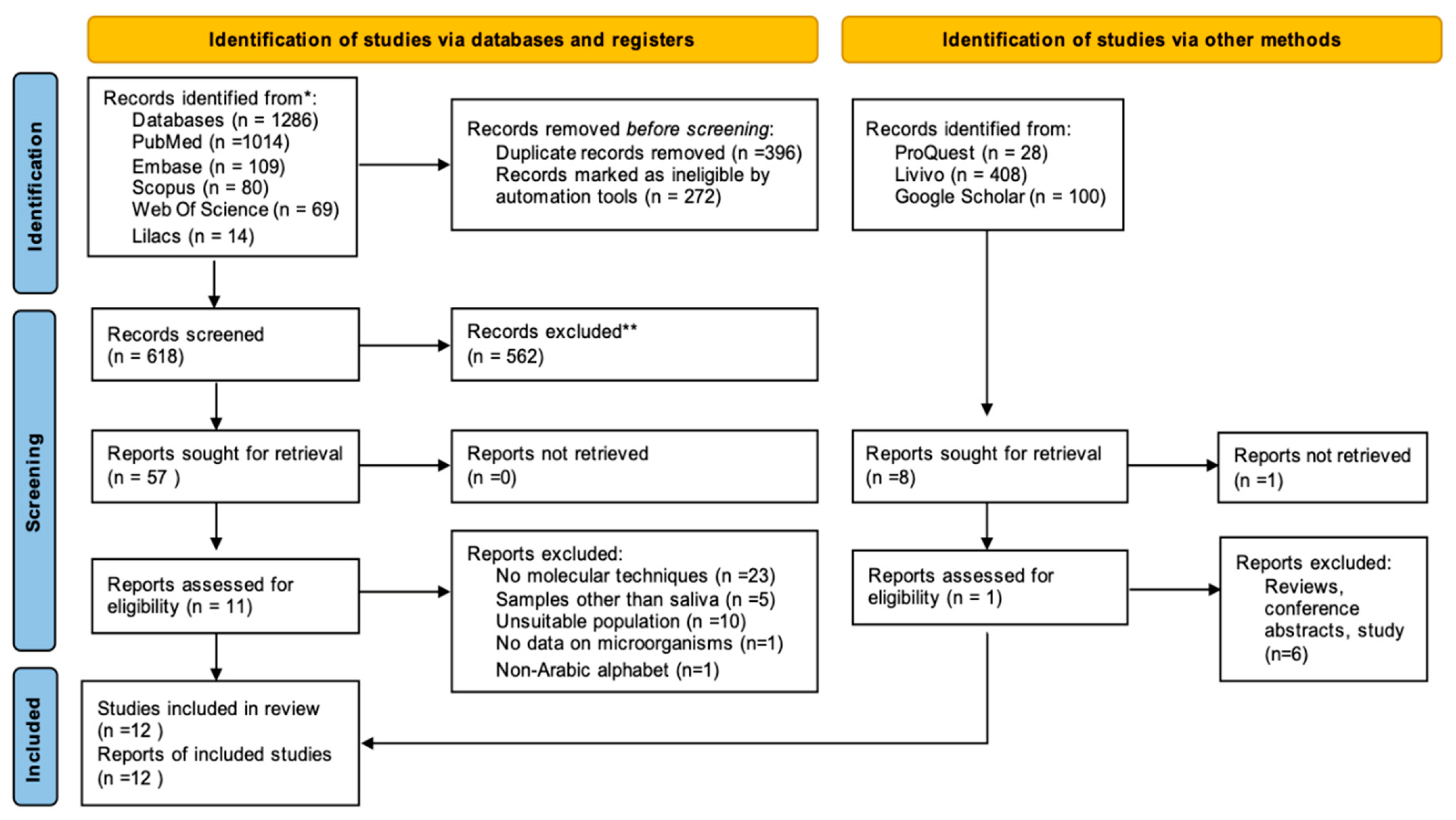
2.3. Studies Selection
2.4. Data Collection and Analysis
2.5. Methodological Quality Assessment
2.6. Certainty of the Evidence
3. Results
3.1. Characteristics of Selected Studies
3.2. Salivary Microbiota
4. Discussion
5. Conclusions
6. Records
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, A.; Ganley, K.; Miecznikowski, J.C.; Haase, E.M.; Barnes, V.; Scannapieco, F.A. The salivary microbiome of diabetic and non-diabetic adults with periodontal disease. J. Periodontol. 2019, 90, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Latti, B.R.; Kalburge, J.V.; Birajdar, S.B.; Latti, R.G. Evaluation of relationship between dental caries, diabetes mellitus and oral microbiota in diabetics. J. Oral Maxillofac. Pathol. JOMFP 2018, 22, 282. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Santos, A.; Martins-Mendes, D.; Gaya-Vidal, M.; Perez-Pardal, L.; Beja-Pereira, A. Characterization of the Oral Microbiome of Medicated Type-2 Diabetes Patients. Front. Microbiol. 2021, 12, 610370. [Google Scholar] [CrossRef]
- Kampoo, K.; Teanpaisan, R.; Ledder, R.G.; McBain, A.J. Oral bacterial communities in individuals with type 2 diabetes who live in southern Thailand. Appl. Environ. Microbiol. 2014, 80, 662–671. [Google Scholar] [CrossRef]
- Grisi, D.C.; Vieira, I.V.; de Almeida Lima, A.K.; de Oliveira Mattos, M.C.; Damé-Teixeira, N.; Salles, L.P.; de Oliveira, L.A.; Stefani, C.; do Carmo Machado Guimarães, M. The complex interrelationship between diabetes mellitus, oral diseases and general health. Curr. Diabetes Rev. 2022, 18, 8–22. [Google Scholar] [CrossRef]
- Graves, D.T.; Correa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef]
- Almusawi, M.A.; Gosadi, I.; Abidia, R.; Almasawi, M.; Khan, H.A. Potential risk factors for dental caries in Type 2 diabetic patients. Int. J. Dent. Hyg. 2018, 16, 467–475. [Google Scholar] [CrossRef]
- Yamashita, J.M.; Moura-Grec, P.G.; Capelari, M.M.; Sales-Peres, A.; Sales-Peres, S.H. Manifestações bucais em pacientes portadores de Diabetes Mellitus: Uma revisão sistemática. Rev. Odontol. UNESP 2013, 42, 211–220. [Google Scholar] [CrossRef]
- Vieira Lima, C.P.; Arlindo Chagas, L.F.; Rego Marques, R.C.; Grisi, D.C.; Salles, L.P.; do Carmo Machado Guimarães, M.; Dame-Teixeira, N. Can hyperglycemia be associated with caries activity and root caries in adults? Prim. Care Diabetes 2022, 17, 48–54. [Google Scholar] [CrossRef]
- Goodson, J.M.; Hartman, M.-L.; Shi, P.; Hasturk, H.; Yaskell, T.; Vargas, J.; Song, X.; Cugini, M.; Barake, R.; Alsmadi, O.; et al. The salivary microbiome is altered in the presence of a high salivary glucose concentration. PLoS ONE 2017, 12, e0170437. [Google Scholar] [CrossRef]
- Marques, R.C.R.; da Silva, J.R.; Lima, C.P.V.; Stefani, C.M.; Damé-Teixeira, N. Salivary parameters of adults with Diabetes Mellitus: A systematic review and meta-analysis: Short title: Salivary parameters in diabetes mellitus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2022, 134, 176–189. [Google Scholar] [CrossRef]
- de Lima, A.K.A.; Amorim dos Santos, J.; Stefani, C.M.; Almeida de Lima, A.d.; Damé-Teixeira, N. Diabetes mellitus and poor glycemic control increase the occurrence of coronal and root caries: A systematic review and meta-analysis. Clin. Oral Investig. 2020, 24, 3801–3812. [Google Scholar] [CrossRef]
- Vieira Lima, C.P.; Grisi, D.C.; Guimarães, M.D.; Salles, L.P.; Kruly, P.D.; Do, T.; Dos Anjos Borges, L.G.; Dame-Teixeira, N. Enrichment of sulphate-reducers and depletion of butyrate-producers may be hyperglycaemia signatures in the diabetic oral microbiome. J. Oral Microbiol. 2022, 14, 2082727. [Google Scholar] [CrossRef]
- Kori, J.A.; Saleem, F.; Ullah, S.; Azim, M.K. Characterization of Oral bacteriome dysbiosis in type 2 diabetic patients. medRxiv 2020. medRxiv:2020-04. [Google Scholar] [CrossRef]
- Casarin, R.C.V.; Barbagallo, A.; Meulman, T.; Santos, V.R.; Sallum, E.A.; Nociti, F.H.; Duarte, P.M.; Casati, M.Z.; Gonçalves, R.B. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal Res. 2013, 48, 30–36. [Google Scholar] [CrossRef]
- Hintao, J.; Teanpaisan, R.; Chongsuvivatwong, V.; Ratarasan, C.; Dahlen, G. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral Microbiol. Immunol. 2007, 22, 175–181. [Google Scholar] [CrossRef]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef]
- Marsh, P.D.; Do, T.; Beighton, D.; Devine, D.A. Influence of saliva on the oral microbiota. Periodontology 2016, 70, 80–92. [Google Scholar] [CrossRef]
- Munn, Z.; Stern, C.; Aromataris, E.; Lockwood, C.; Jordan, Z. What kind of systematic review should I conduct? A proposed typology and guidance for systematic reviewers in the medical and health sciences. BMC Med. Res. Methodol. 2018, 18, 5. [Google Scholar] [CrossRef]
- Moola, S.; Munn, Z.; Tufanaru, C.; Aromataris, E.; Sears, K.; Sfetcu, R.; Currie, M.; Qureshi, R.; Mattis, P.; Lisy, K.; et al. Checklist for analytical cross sectional studies. In Joanna Briggs Institute Reviewer’s Manual; The Joanna Briggs Institute: Adelaide, South Australia, 2017. [Google Scholar]
- Murad, M.H.; Mustafa, R.A.; Schünemann, H.J.; Sultan, S.; Santesso, N. Rating the certainty in evidence in the absence of a single estimate of effect. BMJ Evid.-Based 2017, 22, 85–87. [Google Scholar] [CrossRef]
- Al-Rawi, N.; Al-Marzooq, F. The Relation between Periodontopathogenic Bacterial Levels and Resistin in the Saliva of Obese Type 2 Diabetic Patients. J. Diabetes Res. 2017, 2017, 2643079. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, M.; Xia, L.; Fang, Z.; Yu, S.; Gao, J.; Feng, Q.; Yang, P. Alteration of salivary microbiome in periodontitis with or without type-2 diabetes mellitus and metformin treatment. Sci. Rep. 2020, 10, 15363. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Honda-Ogawa, M.; Ikebe, K.; Notomi, Y.; Iwamoto, Y.; Shirobayashi, I.; Hata, S.; Kibi, M.; Masayasu, S.; Sasaki, S.; et al. Characterizations of oral microbiota in elderly nursing home residents with diabetes. J. Oral Sci. 2017, 59, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, S.; Wang, Y.; Wang, Z.; Ding, W.; Sun, X.; He, K.; Feng, Q.; Zhang, X. Changes of saliva microbiota in the onset and after the treatment of diabetes in patients with periodontitis. Aging 2020, 12, 13090–13114. [Google Scholar] [CrossRef]
- Saeb, A.T.; Al-Rubeaan, K.A.; Aldosary, K.; Raja, G.U.; Mani, B.; Abouelhoda, M.; Tayeb, H.T. Relative reduction of biological and phylogenetic diversity of the oral microbiota of diabetes and pre-diabetes patients. Microb. Pathog. 2019, 128, 215–229. [Google Scholar] [CrossRef]
- Chumponsuk, T.; Gruneck, L.; Gentekaki, E.; Jitprasertwong, P.; Kullawong, N.; Nakayama, J.; Popluechai, S. The salivary microbiota of Thai adults with metabolic disorders and association with diet. Arch. Oral Biol. 2021, 122, 105036. [Google Scholar] [CrossRef]
- Liu, Y.K.; Chen, V.; He, J.Z.; Zheng, X.; Xu, X.; Zhou, X.D. A salivary microbiome-based auxiliary diagnostic model for type 2 diabetes mellitus. Arch. Oral Biol. 2021, 126, 105118. [Google Scholar] [CrossRef]
- Anbalagan, R.; Srikanth, P.; Mani, M.; Barani, R.; Seshadri, K.G.; Janarthanan, R. Next generation sequencing of oral microbiota in Type 2 diabetes mellitus prior to and after neem stick usage and correlation with serum monocyte chemoattractant-1. Diabetes Res. Clin. Pract. 2017, 130, 204–210. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. BioMed Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Colditz, G.; Willett, W.C.; Rotnitzky, A.; Manson, J.E. Weight gain as a risk factor for clinical diabetes mellitus in women. Ann. Intern. Med. 1995, 122, 481–486. [Google Scholar] [CrossRef]
- Nasidze, I.; Li, J.; Quinque, D.; Tang, K.; Stoneking, M.J. Global diversity in the human salivary microbiome. Genome Res. 2009, 19, 636–643. [Google Scholar] [CrossRef]
- Daniel, R.; Gokulanathan, S.; Shanmugasundaram, N.; Lakshmigandhan, M.; Kavin, T. Diabetes and periodontal disease. J. Pharm. Bioallied Sci. 2012, 4 (Suppl. S2), S280. [Google Scholar] [CrossRef]
- Tan, E.C.; Lexomboon, D.; Sandborgh-Englund, G.; Haasum, Y.; Johnell, K. Medications that cause dry mouth as an adverse effect in older people: A systematic review and metaanalysis. J. Am. Geriatr. Soc. 2018, 66, 76–84. [Google Scholar] [CrossRef]
- Hassan, A.; Pratyusha, F. Diabetes and oral diseases—A review. IP J. Nutr. Metab. Health Sci. 2020, 3, 6–9. [Google Scholar] [CrossRef]
- Weng, C.-T.; Huang, S.-L.; Yang, H.-W.; Kao, C.-C.; Wei, C.-C.; Huang, Y.-F. Oral microbiota in xerostomia patients—A preliminary study. J. Dent. Sci. 2022, 17, 324–330. [Google Scholar] [CrossRef]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Nguyen, N.-P.; Warnow, T.; Pop, M.; White, B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes 2016, 2, 16004. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Welch, D.M.; Relman, D.A.; Sogin, M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008, 4, e1000255. [Google Scholar] [CrossRef]
- Mirzayi, C.; Renson, A.; Furlanello, C.; Sansone, S.-A.; Zohra, F.; Elsafoury, S.; Geistlinger, L.; Kasselman, L.J.; Eckenrode, K.; van de Wijgert, J.; et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.; Mulrow, C.D.; Shamseer, L.; Moher, D. Mapping of reporting guidance for systematic reviews and meta-analyses generated a comprehensive item bank for future reporting guidelines. J. Clin. Epidemiol. 2020, 118, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; McKenzie, J.E.; Sowden, A.; Katikireddi, S.V.; Brennan, S.E.; Ellis, S.; Hartmann-Boyce, J.; Ryan, R.; Shepperd, S.; Thomas, J.; et al. Synthesis without meta-analysis (SWiM) in systematic reviews: Reporting guideline. BMJ 2020, 368, l6890. [Google Scholar] [CrossRef] [PubMed]
- Aitken-Saavedra, J.; Lund, R.G.; González, J.; Huenchunao, R.; Perez-Vallespir, I.; Morales-Bozo, I.; Urzúa, B.; Tarquinio, S.C.; Maturana-Ramírez, A.; Martos, J.; et al. Diversity, frequency and antifungal resistance of Candida species in patients with type 2 diabetes mellitus. Acta Odontol Scand. 2018, 76, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Almusawi, M.A.; Gosadi, I.; Abidia, R.; Almasawi, M.; Alrashood, S.T.; Ekhzaimy, A.; Alhomida, A.S.; Khan, H.A. Association between salivary factors and cariogenic bacteria in type-2 diabetes patients. J. King Saud Univ. Sci. 2020, 32, 2617–2621. [Google Scholar] [CrossRef]
- Amato, R.; Pecora, A. “Incidenza della candidosi orale in un gruppo campione di soggetti affetti da diabete mellito” [Incidence of oral candidiasis in a sample group of diabetics]. Boll. Soc. Ital. Biol. Sper. 1983, 59, 532–534. [Google Scholar]
- Baena-Monroy, T.; Moreno-Maldonado, V.; Franco-Martínez, F.; Aldape-Barrios, B.; Quindós, G.; Sánchez-Vargas, L.O. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Buccal 2005, 10 (Suppl. S1), E27–E39. [Google Scholar]
- Kulshrestha, R.; Neral, A.; Srinivasa, T.S.; Baig, S.A. Comparison of Oral Microflora of Diabetic and Non-Diabetic Patients with Periodontitis. J. Pure Appl. Microbio. 2011, 5, 883–886. [Google Scholar]
- Balan, P.; Gogineni, S.B.; Kumari, S.; Shetty, V.; Rangare, A.L.; Castelino, R.L.; Areekat, F. Candida Carriage Rate and Growth Characteristics of Saliva in Diabetes Mellitus Patients: A Case-Control Study. J. Dent. Res. Dent. Clin. Dent. Prospect. 2015, 9, 274–279. [Google Scholar] [CrossRef]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; DeVizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef]
- Bernabe, B.P.; Gilbert, J.A. Dynamic Core Taxonomical and Metabolic Signatures in the Perinatal Period. In Reproductive Sciences; Sage Publications Inc.: Thousand Oaks, CA, USA, 2019; Volume 26. [Google Scholar]
- Bianchi, C.M.; Bianchi, H.A.; Tadano, T.; Depaula, C.R.; Hoffmann-Santos, H.D.; Leite, D.P.; Hahn, R.C. Factors Related to Oral Candidiasis in Elderly Users and Non-Users of Removable Dental Prostheses. Rev. Inst. Med. Trop. Sao Paulo 2016, 58, 17. [Google Scholar] [CrossRef]
- Bremenkamp, R.; Caris, A.; Jorge, A.; Back-Brito, G.; Mota, A.; Balducci, I.; Brighenti, F.; Koga-Ito, C. Prevalence and antifungal resistance profile of Candida spp. oral isolates from patients with type 1 and 2 diabetes mellitus. Arch. Oral Biol. 2011, 56, 549–555. [Google Scholar] [CrossRef]
- Chouhan, S.; Kallianpur, S.; Prabhu, K.; Tijare, M.; Kasetty, S.; Gupta, S. Candidal Prevalence in Diabetics and its Species Identification. Int. J. Appl. Basic Med. Res. 2019, 9, 49–54. [Google Scholar] [CrossRef]
- Crusell, M.K.W.; Brink, L.R.; Nielsen, T.; Allin, K.H.; Hansen, T.; Damm, P.; Lauenborg, J.; Hansen, T.H.; Pedersen, O. Gestational diabetes and the human salivary microbiota: A longitudinal study during pregnancy and postpartum. BMC Pregnancy Childbirth 2020, 20, 69. [Google Scholar] [CrossRef]
- Darwazeh, A.M.G.; MacFarlane, T.W.; McCuish, A.; Lamey, P.-J. Mixed salivary glucose levels and candidal carriage in patients with diabetes mellitus. J. Oral Pathol. Med. 1991, 20, 280–283. [Google Scholar] [CrossRef]
- Davila-Perez, C.; Amano, A.; Alpuche-Solis, A.; Patiño-Marin, N.; Pontigo, A.P.; Hamada, S.; Loyola-Rodriguez, J.P. Distribution of genotypes of Porphyromonas gingivalis in type 2 diabetic patients with periodontitis in Mexico. J. Clin. Periodontol. 2007, 34, 25–30. [Google Scholar] [CrossRef]
- Farizal, J.; Dewa, E.A.R.S. Identifikasi Candida Albican Pada Saliva Wanita Penderita Diabetes Melitus. J. Teknol. Lab. 2017, 6, 67–74. [Google Scholar] [CrossRef]
- Heggendorn, F.L.; Gonçalves, L.; Dias, E.P.; Junior, A.S.; Galvão, M.M.; Lutterbach, M.T.S. Detection of sulphate-reducing bacteria in human saliva. Acta Odontol. Scand. 2013, 71, 1458–1463. [Google Scholar] [CrossRef]
- Janem, W.F.; Scannapieco, F.A.; Sabharwal, A.; Tsompana, M.; Berman, H.A.; Haase, E.M.; Miecznikowski, J.C.; Mastrandrea, L.D. Salivary inflammatory markers and microbiome in normoglycemic lean and obese children compared to obese children with type 2 diabetes. PLoS ONE 2017, 12, e0172647. [Google Scholar] [CrossRef]
- Javed, F.; Klingspor, L.; Sundin, U.; Altamash, M.; Klinge, B.; Engström, P.-E. Periodontal conditions, oral Candida albicans and salivary proteins in type 2 diabetic subjects with emphasis on gender. BMC Oral Health 2009, 9, 12. [Google Scholar] [CrossRef]
- Khovidhunkit, S.O.; Suwantuntula, T.; Thaweboon, S.; Mitrirattanakul, S.; Chomkhakhai, U.; Khovidhunkit, W. Xerostomia, hyposalivation, and oral microbiota in type 2 diabetic patients: A preliminary study. J. Med. Assoc. Thail. Chotmaihet Thangphaet 2009, 92, 1220–1228. [Google Scholar]
- Liao, Y.T.; He, L.; Meng, H.X.; Li, P.; Sha, Y.Q.; Wang, X.Y. Detection of periodontal pathogens from saliva of type 2 diabetic patients in urban area of Beijing. Zhonghua Kou Qiang Yi Xue Za Zhi=Zhonghua Kouqiang Yixue Zazhi Chin. J. Stomatol. 2013, 48, 144–149. [Google Scholar]
- Lira-Junior, R.; Åkerman, S.; Klinge, B.; Boström, E.A.; Gustafsson, A. Salivary microbial profiles in relation to age, periodontal, and systemic diseases. PLoS ONE 2018, 13, e0189374. [Google Scholar] [CrossRef]
- Minty, M.; Loubières, P.; Canceill, T.; Azalbert, V.; Burcelin, R.; Tercé, F.; Blasco-Baque, V. Gender-associated differences in oral microbiota and salivary biochemical parameters in response to feeding. J. Physiol. Biochem. 2021, 77, 155–166. [Google Scholar] [PubMed]
- Mizushiri, S.; Daimon, M.; Murakami, H.; Murabayashi, M.; Matsuhashi, Y.; Kamba, A. 1555-P: Differences in Oral Microbiota as Risk Factors for Periodontal Disease in Diabetic Subjects. Diabetes 2019, 68, 1555-P. [Google Scholar] [CrossRef]
- Nabee, Z.; Jeewon, R.; Pugo-Gunsam, P. Oral dysbacteriosis in type 2 diabetes and its role in the progression to cardiovascular disease. Afr. Health Sci. 2017, 17, 1082–1091. [Google Scholar] [CrossRef]
- Naik, R. Assessing oral candidal carriage with mixed salivary glucose levels as non-invasive diagnostic tool in type-2 diabetics of davangere, karnataka, India. J. Clin. Diagn. Res. JCDR 2014, 8, ZC69–ZC72. [Google Scholar] [CrossRef]
- Omori, M.; Kato-Kogoe, N.; Sakaguchi, S.; Kamiya, K.; Fukui, N.; Gu, Y.-H.; Nakamura, S.; Nakano, T.; Hoshiga, M.; Imagawa, A.; et al. Characterization of salivary microbiota in elderly patients with type 2 diabetes mellitus: A matched case-control study. Clin. Oral Investig. 2022, 26, 493–504. [Google Scholar] [CrossRef]
- Popova, P.; Tikhonov, E.; Pinto, Y.; Tkachuk, A.; Vasukova, E.; Dronova, A.; Bolotko, Y.; Frishman, S.; Pustozerov, E.; Pervunina, T.; et al. The Influence of Gestational Diabetes Treatment Modalities on Gut and Saliva Microbiome Composition; European Association for the Study of Diabetes: Montecatini Terme, Italy, 2020. [Google Scholar]
- Radhakrishnan, P.; Anbalagan, R.; Barani, R.; Mani, M.; Seshadri, K.G.; Srikanth, P. Sequencing of Porphyromonas gingivalis from saliva in patients with periodontitis and type 2 diabetes mellitus. Indian J. Med. Microbiol. 2019, 37, 54–59. [Google Scholar] [CrossRef]
- Rajakumari, M.L.; Kumari, P.S. Isolation, identification and characterization of bacteria occurring in the oral cavity of diabetic and non diabetic individuals. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 1294–1301. [Google Scholar]
- Salminen, A.; Kopra, K.A.E.; Hyvärinen, K.; Paju, S.; Mäntylä, P.; Buhlin, K.; Nieminen, M.S.; Sinisalo, J.; Pussinen, P. Quantitative PCR analysis of salivary pathogen burden in periodontitis. Front. Cell. Infect. Microbiol. 2015, 5, 69. [Google Scholar] [CrossRef]
- Salminen, A.; Gursoy, U.K.; Paju, S.; Hyvärinen, K.; Mäntylä, P.; Buhlin, K.; Könönen, E.; Nieminen, M.S.; Sorsa, T.; Sinisalo, J.; et al. Salivary biomarkers of bacterial burden, inflammatory response, and tissue destruction in periodontitis. J. Clin. Periodontol. 2014, 41, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Samnieng, P.; Sonthayasathapon, S.; Siriwat, M.; Jeamanukulkit, S. A cross-sectional study of the Candidal Species Isolated in the Oral Cavities of Type II Diabetic Patients. Makara J. Health Res. 2017, 21, 99–103. [Google Scholar] [CrossRef]
- Shanker, J.; Setty, P.; Arvind, P.; Nair, J.; Bhasker, D.; Balakrishna, G.; Kakkar, V.V. Relationship between periodontal disease, Porphyromonas gingivalis, peripheral vascular resistance markers and coronary artery disease in Asian Indians. Thromb. Res. 2013, 132, e8–e14. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, M.P.; Puranik, R.S.; Vanaki, S.S.; Puranik, S.R.; Shetty, P.; Shenoy, R. A comparative study of oral candidal species carriage in patients with type1 and type2 diabetes mellitus. J. Oral Maxillofac. Pathol. JOMFP 2014, 18 (Suppl. S1), S60–S65. [Google Scholar] [CrossRef]
- Soni, A.P.; Astekar, M.; Metgud, R.; Vyas, A.; Ramesh, G.; Sharma, A.; Verma, M. Candidal carriage in diabetic patients: A microbiological study. J. Exp. Ther. Oncol. 2019, 13, 15–21. [Google Scholar]
- Suárez, B.L.; Álvarez, M.I.; de Bernal, M.; Collazos, A. Candida species and other yeasts in the oral cavities of type 2 diabetic patients in Cali, Colombia. Colomb. Med. 2013, 44, 26–30. [Google Scholar] [CrossRef]
- Tatarakis, N.; Kinney, J.S.; Inglehart, M.; Braun, T.M.; Shelburne, C.; Lang, N.P.; Giannobile, W.V.; Oh, T.-J. Clinical, microbiological, and salivary biomarker profiles of dental implant patients with type 2 diabetes. Clin. Oral Implant. Res. 2014, 25, 803–812. [Google Scholar] [CrossRef]
- Vijayalakshmi, L.; Raj, J.B.S.; Kavitha, J.; Krishnaraj, S.; Manovijay, B.; Manikandan, D. A Case-Control Study to Evaluate Candidal Parameters in the Oral Cavity of Patients with Type 2 Diabetes Mellitus. J. Pharm. Bioallied Sci. 2020, 12 (Suppl. S1), S389–S393. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y.; et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef]
- Wang, R.-R.; Xu, Y.-S.; Ji, M.-M.; Zhang, L.; Li, D.; Lang, Q.; Zhang, L.; Ji, G.; Liu, B.-C. Association of the oral microbiome with the progression of impaired fasting glucose in a Chinese elderly population. J. Oral Microbiol. 2019, 11, 1605789. [Google Scholar] [CrossRef]
- Wei, Y.-S.; Hsiao, Y.-C.; Su, G.-W.; Chang, Y.-R.; Lin, H.-P.; Wang, Y.-S.; Tsai, Y.-T.; Liao, E.-C.; Chen, H.-Y.; Chou, H.-C.; et al. Identification of hyperglycemia-associated microbiota alterations in saliva and gingival sulcus. Arch. Biochem. Biophys. 2020, 682, 108278. [Google Scholar] [CrossRef]
| Study | Mq * | Group with DM | Controls | Salivary Collection | Microbiota Assessment |
|---|---|---|---|---|---|
| Almeida-Santos, 2021 Portugal | − | T2D reported and treated for more than 14 years (HbA1c = 7.2% ± 1.0) n = 25, mean age: 62.7 ± 7.0 yo. | Those reporting no T2D n = 25, mean age: 60.0 ± 8.8 yo. | Not reported ** | 16S rRNA V3–V4 region |
| Al-Rawi, 2017 United Arab Emirates | − | Obese with T2D (FBG = 205.5 ± 83.9 mg/dL) n = 26, mean age: 51.1 ± 5.7 years Obese without T2D (FBG = 106.2 ± 24.9 mg/dL) n = 26, mean age: 47.9 ± 5.7 yo. | Non-obese and non-diabetic (FBG = 94 ± 17.1 mg/dL) n = 26, mean age: 47.4 ± 5 yo. | Whole saliva, unstimulated | RT-PCR using taxon-specific primers targeting six proteolytic bacteria, and including a universal 16S rRNA primer |
| Anbalagan, 2017 India | − | T2D reported (HbA1c = 11.33% ± 1.56) n = 24, mean age: 50.5 ± 14.72 yo. | Those reporting no T2D (HbA1C < 6.5%) n = 10 | Whole saliva, unstimulated | 16S rRNA V6 region |
| Chumponsuk, 2021 Thailand | + | T2D according to FBG (FBG= 119.37 ± 37.12 mg/dL) n = 25, mean age: 50.64 ± 13.75 yo. | Non-diabetics were categorized into 3 groups according to BMI (FBG = 96.11 ± 6.7 mg/dL) n = 80, mean age: 44.82 ± 2.5 yo. | Whole saliva, unstimulated | RT-PCR using several taxon-specific primers targeting 16S rRNA |
| Kori, 2020 Pakistan | − | T2D reported (HbA1c = 8.3% ± 1.7) n = 49, mean age: 53.1± 7.9 yo. | Those reporting no T2D (HbA1c= 5.2% ± 0.4) n = 50, mean age: 39.7 ± 11.8 yo. | Whole saliva, unstimulated | 16S rRNA V3-V4 region |
| Vieira Lima, 2022 Brazil | ++ | T2D reported and treated (HbA1c = 8.83% ± 2.02, FBG= 148.13 ± 65.43 mg/dL) n = 23, mean age: 58.52 ± 8.5 yo. | Those reporting no T2D (HbA1c = 5.25% ± 0.41, FBG = 89.67 ± 13.07 mg/dL) n = 25, mean age: 43.13 ± 12.98 yo. | Whole saliva, unstimulated and stimulated | 16S rRNA V4 region |
| Liu, 2021 China | + | Reported and untreated T2D (HbA1c = 8.51% ± 2.33) n = 24, mean age: 47(33–65) yo. | Those reporting no T2D (HbA1c = 5.25% ± 0.41) n = 21, mean age: 47.24(35∼61) yo. | Whole saliva, unstimulated | 16S rRNA V3-V4 region |
| Ogawa, 2017 Japan | − | Those who reported treatment for T2D (HbA1c = 8.51% ± 2.33) n = 3, mean age: 85.3 ± 4.5 yo. | Those reporting no T2D n = 12, mean age: 83.9 ± 8.4 yo. | Unstimulated saliva | 16S rRNA V4 region |
| Sabharwal, 2018 United States | − | Self-reported T2D (HbA1c = 7.37% ± 0.30) n = 79, mean age: 52.9 ± 2.13 yo. | Those reporting no T2D n = 64, mean age: 39.3 ± 8.5 yo. | Unstimulated saliva | 16S rRNA V3-V4 region |
| Saeb,2019 Saudi Arabia | ++ | T2D reported and treated (HbA1c = 9.5% ± 0.83) for more than 11 years n = 15, mean age: 51.2 ± 3.14 yo. | Those reporting no T2D n = 19, mean age: 41–56 yo. | Whole saliva spat out after tongue rubbing (OM501 Kit for Microbial DNA Analysis) | 16S rRNA qPCR V2-4 and V6-9 regions |
| Sun, 2020 China | ++ | Reported and treated T2D (HbA1c = 7.7% ± 2.06) n = 75, mean age: 54.96 ± 8.95 yo. | Those with no reported T2D (HbA1c = 4.96% ± 0.15) n = 58, mean age: 36.66 ± 11.15 yo. | Unstimulated saliva, after rinsing with tap water | 16S rRNA qPCR V3-V4 region |
| Yang, 2020 China | + | T2D reported or reported and treated (FBG = 11.58 ± 0.99 mmol/L) n = 70, mean age: 54.62 ± 12.13 yo. | Those with no reported T2D (FBG = 5.38 ± 0.48 mmol/L) n = 32, mean age: 49.18 ± 8.72 yo. | Unstimulated saliva, after rinsing with sterile distilled water | 16S rRNA V3-V4 region |
| Microorganism ** | Al-Rawi l, 2017 | Sun, 2020 | Anbalagan, 2017 | Yang, 2020 | Kori, 2020 | Chumponsuk,2021 | Almeida-Santos,2021 | Liu, 2021 | Ogawa, 2017 | Sabharwal, 2019 | Saeb, 2019 | Vieira Lima, 2022 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phyla in general | ||||||||||||
| Firmicutes | ↑ | ↑ | NS | NS | ↑ * | NS | ||||||
| Bacterioidetes | ↑ * | ↑ | NS | NS | ↓ * | NS | ||||||
| Actinobacteria | NS | NS | NS | ↑ * | NS | ↓ * | ||||||
| Proteobacteria | ↓ | ↓ | NS | NS | ↓ * | ↓ | ||||||
| Spirochaetota | NS | NS | ||||||||||
| Euryarchaeota | ||||||||||||
| Fusobacteria | ↑ | NS | NS | |||||||||
| Lactic acid bacteria (LAB) | ||||||||||||
| Streptococcus | ↓ * | ↑ * | NS | NS | NS | |||||||
| Lactobacillus spp. | ↑ * | ↑ | NS | NS | NS | |||||||
| Bifidobacteriaceae, Bifidobacterium | NS | ↑ * | NS | NS | ||||||||
| Scardovia, Parascardovia | ↑ * | NS | ||||||||||
| Enterococcus | ||||||||||||
| Other LAB (Lactococcus, Leuconostoc, Pediococcus, Vagococcus, Aerococcus, Tetragenecoccus, Carnobacterium) | ||||||||||||
| Bacteria producing butyrate or other short chain fatty acids (succinate, acetate, propionate) | ||||||||||||
| Butyvibrio | ↓ | |||||||||||
| Bacteroides | NS | NS | ||||||||||
| Prevotella | ↑ * | ↑ | ↑ | NS | ↓ * | NS | ||||||
| Alloprevotella | ↓ * | |||||||||||
| Paraprevotella | ||||||||||||
| Fusobacterium | ↑ | ↑ * | NS | ↑ * | ||||||||
| Tannerella, T. forsythia | ↑ | ↑ * | ↑ | |||||||||
| Treponema, T. denticola | NS | ↓ * | ||||||||||
| Acid-associated microorganisms (lactate metabolizers, other acidurics) | ||||||||||||
| Veillonella | ↑ * | ↑ | NS | ↑ * | NS | NS | ||||||
| Certainty Assessment | Impact | Certainty | ||||||
|---|---|---|---|---|---|---|---|---|
| № of Studies | Study Design | Risk of Bias | Inconsistency | Indirectness | Imprecision | Other Considerations | ||
| Firmicutes | ||||||||
| 12 | Observational | Not serious a | Very serious b | Not serious | Serious | None | The evidence is very uncertain about the enrichment of Firmicutes in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Proteobacteria | ||||||||
| 12 | Observational | Not serious a | Very serious b | Not serious | Serious c | None | The evidence is very uncertain about the depletion of Proteobacteria in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Prevotella | ||||||||
| 12 | Observational | Not serious a | Very serious b | Not serious | Serious c | None | The evidence is very uncertain about the enrichment of Prevotella in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Fusobacterium | ||||||||
| 12 | Observational studies | Not serious a | Very serious b | Not serious | Serious c | None | The evidence is very uncertain about the enrichment of Fusobacterium in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Veillonella | ||||||||
| 12 | Observational studies | Not serious a | Very serious b | Not serious | Serious c | None | The evidence is very uncertain about the enrichment of Veillonella in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Lactobacillus spp. | ||||||||
| 12 | Observational | Not serious a | Very serious b | Not serious | Serious c | None | The evidence is very uncertain about the enrichment of Lactobacillus spp. in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D. | ⨁◯◯◯ Very low |
| Tannerella/T. forsythia | ||||||||
| 12 | Observational | Not serious a | Serious b | Not serious | Serious c | None | Tannerella/T. forsythia may be enriched in the saliva of adults with T2D when compared to individuals without T2D or with controlled T2D, although the certainty is low. | ◯⨁◯◯ Low |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cena, J.A.; Reis, L.G.; de Lima, A.K.A.; Vieira Lima, C.P.; Stefani, C.M.; Dame-Teixeira, N. Enrichment of Acid-Associated Microbiota in the Saliva of Type 2 Diabetes Mellitus Adults: A Systematic Review. Pathogens 2023, 12, 404. https://doi.org/10.3390/pathogens12030404
Cena JA, Reis LG, de Lima AKA, Vieira Lima CP, Stefani CM, Dame-Teixeira N. Enrichment of Acid-Associated Microbiota in the Saliva of Type 2 Diabetes Mellitus Adults: A Systematic Review. Pathogens. 2023; 12(3):404. https://doi.org/10.3390/pathogens12030404
Chicago/Turabian StyleCena, Jéssica Alves, Letícia Gonçalves Reis, Ana Karolina Almeida de Lima, Camilla Pedrosa Vieira Lima, Cristine Miron Stefani, and Naile Dame-Teixeira. 2023. "Enrichment of Acid-Associated Microbiota in the Saliva of Type 2 Diabetes Mellitus Adults: A Systematic Review" Pathogens 12, no. 3: 404. https://doi.org/10.3390/pathogens12030404
APA StyleCena, J. A., Reis, L. G., de Lima, A. K. A., Vieira Lima, C. P., Stefani, C. M., & Dame-Teixeira, N. (2023). Enrichment of Acid-Associated Microbiota in the Saliva of Type 2 Diabetes Mellitus Adults: A Systematic Review. Pathogens, 12(3), 404. https://doi.org/10.3390/pathogens12030404





