Abstract
Salmonellosis remains the second most common zoonosis in Europe. Resistance to fluoroquinolones (FQs) in Salmonella has been increasing worldwide, with WHO considering FQ-resistant Salmonella spp. as high-priority pathogens. The aim of this study was a retrospective analysis of the molecular mechanisms of FQ resistance, detected among clinical ciprofloxacin-resistant Salmonella enterica belonging to the most common serotypes. The whole genome sequences (WGS) of tested isolates were also analysed for the occurrence of other antimicrobial resistance determinants. Out of a total of 1051 Salmonella collected in the years 2018–2019, 447 strains belonging to the most common serotypes in Poland were selected were screened for FQ resistance using the pefloxacin disc test according to EUCAST recommendations. All pefloxacin-resistant isolates were confirmed as ciprofloxacin-resistant using the E-test. A total of 168 (37.6%) Salmonella enterica, which belonged to seven serotypes, were resistant to ciprofloxacin (mostly Hadar, Virchow and Newport). A hundred randomly selected Salmonella were investigated by WGS. A total of 127 QRDR mutations in GyrA and ParC were identified in 93 isolates. The qnr genes were the only PMQR determinants detected and were found in 19% of the sequenced isolates. Moreover, 19 additional resistance genes (including: bla,, tet, sul, aad, aac-, ant-, aph-, floR, cmlA) were identified among the FQ-resistant Salmonella tested that confer resistance to clinically important antibiotics such as β-lactams, tetracyclines, sulphonamides, aminoglycosides and phenicol, respectively). In conclusion, FQ resistance of human Salmonella in Poland is rising towards a critical level and needs to be tightly monitored.
1. Introduction
Non-typhoidal Salmonella (NTS), including Salmonella enterica subspecies enterica, which is subdivided into more than 2500 serovars based on antigenic differences in the lipopolysaccharide O antigen and two flagellin structures, is a frequent human pathogen [1]. Salmonellosis as a foodborne disease with a wide range of hosts still remains a global public health challenge. In Europe, salmonellosis is the second most common zoonosis in humans, with 87,923 confirmed human cases, as well as the most frequent cause of foodborne outbreaks accounting for 17.9% of all foodborne outbreaks reported in 2019 [2]. In the USA alone, approximately 1.2 million human cases, with 450 fatal cases, occur each year due to Salmonella infection [3]. Generally, NTS infections are transmitted to humans via the consumption of contaminated water and food, particularly animal-derived products. In most cases, NTS infection causes self-limiting diarrhoea. However, a life-threatening systemic infection may occur, in which case, antimicrobial treatment is required, especially in high-risk groups, such as elderly or immunocompromised patients [3]. Recently, antimicrobial resistance (AMR) among Salmonella human isolates, including resistance to fluoroquinolones and the third generation of cephalosporins that are the current drugs of choice for salmonellosis treatment, have been globally encountered with high frequency, also in Europe, making the empiric treatment of salmonellosis more challenging [2,4].
Although, in 2016, the World Health Organisation (WHO) listed fluoroquinolones (FQ) as a critically important class of antibacterial drugs [5], FQs have been widely used to treat various infections, including invasive non-typhoidal Salmonella infections in adults. Furthermore, FQs have also been used in animals to prevent, control and treat infections [6]. The widespread use of FQ to treat both humans and animals creates a selective pressure that promotes the horizontal transfer of resistance genes and the development of antimicrobial resistance, leading to the selection of resistant bacterial clones among pathogenic, commensal or even environmental bacteria [2]. FQ resistance among Salmonella has been on the rise worldwide for the past two decades [2,7,8,9]. In 2017, the WHO considered FQ-resistant Salmonella spp. as pathogens for which novel antibiotics are urgently required [10].
FQs’ main targets are two enzymes essential for bacterial replication: DNA gyrase and topoisomerase IV. In Enterobacterales, including Salmonella, quinolone resistance typically develops from the accumulation of chromosomal mutations in the quinolone resistance-determining region (QRDR) of genes encoding the subunits of DNA gyrase (GyrA, GyrB) and topoisomerase IV (ParC, ParE), primarily in GyrA and ParC [8,11,12]. Additionally, since the late 1990s, three types of plasmid-mediated quinolone resistance (PMQR) mechanisms have been identified: Qnr, which protects target enzymes; AAC(6’)-Ib-cr, which mediates the acetylation of certain quinolones; QepA and OqxAB, which produce mobile efflux pumps [13]. Although PMQRs alone are considered factors that can provide only a low level of resistance, their presence may aid bacteria to survive under low FQ pressure sufficiently to develop mutations in the DNA gyrase and/or topoisomerase IV genes, and to acquire high-level resistance to FQs [14]. These molecular mechanisms can be accumulative and together influence the observed FQ minimum inhibitory concentration (MIC) value. The genotypic features of ciprofloxacin non-susceptibility caused by the QRDR mutations and the PMQR mechanisms among NTS human isolates can vary in time and countries, but most frequently are attributable to the GyrA mutations in 83 and/or 87 codon and/or the ParC substitution in 80 codon and PMQR presence, mainly Qnr [14].
In Poland, generally, about 8000 confirmed salmonellosis cases are detected annually (ranging from 7315 in 2013 to 9718 in 2016) [15]. Furthermore, there is a lack of specific information on AMR incidence in Salmonella because of the insufficient number of isolates tested for phenotypic and genetic antimicrobial resistance by Polish health authorities. Therefore, it is difficult to understand the real level of antimicrobial resistance in Salmonella. Until now, only limited data have been available with regard to the distribution and molecular characteristics of FQ-resistant Salmonella enterica isolated from humans. Recently, we reported on the antimicrobial resistance of S. Kentucky ST 198 [16], one of the most resistant serovars occurring in humans and animals, respectively. Thus, the objective of this study was to investigate the molecular mechanisms of FQ resistance, including mutations in the QRDR (gyrA, gyrB and parC, parE) and PMQR determinants (qnrA, qnrB, qnrS, qnrC, qnrD, aac(6’)-Ib-cr, qepA and oqxAB) in clinical ciprofloxacin-resistant Salmonella enterica belonging to the most prevalent serotypes in Poland. Additionally, genome sequences of the tested ciprofloxacin-resistant Salmonella enterica isolates determined by WGS were also analysed for the presence of other AMR determinants (including: bla,, tet, sul, aad, aac-, ant-, aph-, floR, cmlA that confer resistance to clinically important antibiotics as β-lactams, tetracyclines, sulphonamides, aminoglycosides and phenicol, respectively).
2. Materials and Methods
2.1. Salmonella enterica Isolates and Fluoroquinolone Susceptibility Testing
A total of 1051 Salmonella enterica strains encompassing 43 serotypes isolated from humans (patients, people from contact, convalescents and carriers) were collected in the National Institute of Public Health National Institute of Health–National Research Institute, Warsaw, Poland, in the years 2018–2019. The strains were collected as part of the National Health Program task 2016–2020 “Program for verification of accuracy of human Salmonella strains serotyping in Poland.” The Salmonella strains were identified previously by 14 Provincial Sanitary and Epidemiological Stations in Poland. All the isolates tested were re-serotyped in our laboratory by slide-agglutination methods according to the White–Kauffmann–Le Minor scheme [1] using commercial sera manufactured by: Statens Serum Institut (Denmark), Biomed (Poland) and Immunolab (Poland). Out of 1051 Salmonella enterica collected, 447 strains belonging to the most common serotypes in Poland were selected for FQ resistance screening tests (133 strains from 2018 and 314 strains from 2019). The isolates tested belonged to the following serotypes: Enteritidis (n = 222), Typhimurium (n = 85), Infantis (n = 58), monophasic Typhimurium 1,4,[5],12:i:- (n = 36), Kentucky (n = 18) published previously [16], Hadar (n = 13), Agona (n = 4), Newport (n = 7) and Virchow (n = 4). Pefloxacin (5 µg) screening test was the first criterion for selecting a strain for further study. All the pefloxacin-resistant Salmonella strains detected (zone diameter < 24 mm in accordance with EUCAST criteria; (http://eucast.org (accessed on 5 December 2022)) were confirmed to be resistant to ciprofloxacin by the MIC determination using the E-test strips with an antibiotic concentration gradient (bioMerieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. The MIC results for ciprofloxacin were interpreted using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://eucast.org, accessed on 5 December 2022) criteria, while isolates with MIC > 0.06 mg/L were classified as resistant to ciprofloxacin.
2.2. Whole-Genome Sequencing Analysis
One hundred Salmonella enterica strains were randomly selected for WGS belonged to the Enteritidis (n = 32), Typhimurium (n = 12), Infantis (n = 30), 1,4,[5],12:i:- (n = 6), Hadar (n = 12), Newport (n = 5) and Virchow (n = 3) serotypes with ciprofloxacin MIC > 0.06 mg/L were subjected to further investigation using the whole genome sequence technique. Genomic DNA was isolated from overnight culture in LB based on a manual in-house procedure with the GTC (Guanidine thiocyanate), phenol extraction and precipitation step. The isolated DNA samples were quantified using BioSpectrometer (Eppendorf, Hamburg, Germany). The libraries were constructed using the Illumina DNA Prep chemistry (Illumin, San Diego, CA, USA), and then sequenced on MiSeq (Illumina) using MiSeq Reagent Kit v3.
The raw reads were assembled using CLC Genomics Workbench 22 (Qiagen, Hilden, Germany). Salmonella serotypes were confirmed using the SeqSero tool (CGE) [17]. MLST was assigned based on the raw reads using the MLST 2.0 tool (CGE; database version 22 March 2021) [18].
The acquired antimicrobial resistance genes and chromosomal point mutations were assigned from the raw reads using ResFinder 4.1 and PointFinder (CGE; database from 3 May 2022) [19]. Additionally, all the sequences were submitted to Enterobase, and the results were confirmed using an automatic analytical pipeline. The cgMLST and wgMLST (Multilocus Sequence Typing based on core and whole genome loci) variants were assigned, and specific MLST dendrograms were built. A whole genome SNP analysis was additionally prepared for each Salmonella serotype, and dendrograms were created using the CSI Phylogeny tool [20].
The results and dendrograms were transferred to Microreact (www.microreact.org, accessed on 5 December 2022), where a proper project was established and is available at: https://microreact.org/project/fMudrjXLMz9vzs8tDg5vK1-npz-salmonella-2018-19-publikacja, accessed on 5 December 2022.
3. Results
3.1. Salmonella Strains and Phenotypic Resistance to Fluoroquinolones
A total of 168 (37.6%) Salmonella enterica isolates resistant to ciprofloxacin were obtained after screening with pefloxacin (5 µg) disc diffusion method out of 447 human isolates–133 (39.8%) in 2018 and 314 (41.4%) in 2019, respectively. The FQ-resistant Salmonella strains were sent from different regions of Poland and belonged to seven serotypes, as shown in Figure 1. The number of tested Salmonella strains, the percentage of resistant strains within each serotype and the ciprofloxacin MIC range among the seven serotypes identified in our set of ciprofloxacin-resistant isolates (n = 168) are shown in Table 1.
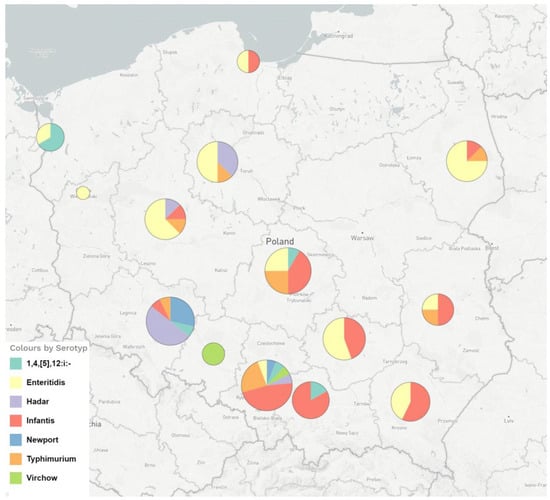
Figure 1.
The geographical distribution and serotypes of FQ-resistant Salmonella enterica strains isolated from humans in Poland in the years 2018–2019 and analysed by WGS.

Table 1.
The number of tested Salmonella serotypes, the number of ciprofloxacin-resistant strains and the ciprofloxacin MIC range for strains isolated from humans in Poland.
The highest percentage of ciprofloxacin-resistant strains was found among Salmonella Hadar, S. Virchow and S. Newport. An equally high percentage of resistant strains was found in S. Infantis and S. Enteritidis.
3.2. Genotypic Antimicrobial Analysis In Silico
3.2.1. Resistance to Fluoroquinolones–QRDR Mutations and PMQR Genes
All the whole genome sequences obtained were entered in Enterobase under accession numbers SAL_LB2507AA–SAL_LB2553AA, SAL_LB2714AA, SAL_LB2752AA–SAL_LB2786AA, SAL_LB2788AA–SAL_LB2793AA and SAL_LB3141–SAL_LB3158AA.
The summary of the WGS results and antimicrobial resistance genes detected among ciprofloxacin-resistant Salmonella enterica strains is shown in Table 2. Additionally, Tables S1–S7 in the Supplementary Materials provide such information as the year of isolation, patient status, ciprofloxacin MIC value, and WGS results including MLST type and resistance genotype for each human isolate tested.

Table 2.
The total number of QRDR mutations and antimicrobial resistance genes including PMQR among the 100 clinical ciprofloxacin-resistant Salmonella enterica isolates of 7 serotypes.
In 93 isolates of 100 Salmonella enterica analysed by means of WGS, a total of 127 QRDR mutations were identified, including 80 mutations in GyrA and 47 in ParC. Only silent mutations were found at the QRDR region of gyrB and parE. The most commonly detected mutations in GyrA were Ser83→Tyr, followed by Asp87→Tyr and Asp87→Asn. In the ParC subunit, substitution was only identified in one codon 57 (Thr→Ser), which was detected in 47 Salmonella isolates (Table 2).
Sixty (60%) of one hundred Salmonella isolates analysed by WGS carried at least one substitution in GyrA or ParC (Tables S1–S7 in the Supplementary Materials). Thirteen isolates (nine S. Hadar and four S. Newport, respectively), had amino acid substitutions in the parC QRDR only. Two substitutions (in GyrA and ParC) were detected in three S. Hadar, thirty S. Infantis and one S. Newport. The distribution of the GyrA and ParC substitutions and PMQR determinants among the seven serotypes of ciprofloxacin-resistant Salmonella enterica isolates are shown in Table 3. No substitution was found in six Salmonella isolates (Typhimurium, n = 1; 1,4,[5],12:i:-, n = 4; Enteritidis, n = 1) that carried qnrB only (Tables S1–S7 in the Supplementary Materials).

Table 3.
The distribution of the GyrA and ParC substitutions and PMQR determinants among the 7 serotypes of ciprofloxacin-resistant Salmonella enterica isolates.
The PMQR determinants were detected in 19 (19%) of 100 Salmonella enterica isolates. The qnr genes were the only PMQR determinant detected. The qnrB19 variant was identified more frequently (in 17 isolates) than other variants: qnrB36 (n = 7); qnrB82 (n = 4) and qnrB67 (n = 2) (Table 2 and Table 3). Three and one of S. Hadar isolates carried a combination of three and four qnrB gene variants, respectively (Table 3 and Table S3). Moreover, 7 of 12 S. Hadar isolates and 4 of 5 S. Newport carried the qnrB gene variants in different combinations and an amino acid substitution in 57 codons of ParC but did not contain any substitution in 83 and/or 87 codons of GyrA. Two isolates of S. Infantis carried qnrS1. Other PMQR determinants, such as qnrA, qnrC, qnrD, aac(6’)-Ib-cr, qepA and oqxAB, were not detected among the Salmonella enterica isolates tested.
3.2.2. In Silico Antimicrobial Resistance Genes Detected among FQ-Resistant Salmonella Isolates
The WGS results for Salmonella isolates resistant to fluoroquinolones are shown in the Supplementary Materials, in Tables S1–S7. In addition to the PMQRs, a total of 19 resistance genes were identified among the FQ-resistant Salmonella tested. These genes were predicted to confer resistance to clinically important classes of antibiotics other than FQs (Table 2 and Tables S1–S7). A list of the resistance genes and their prevalence among Salmonella isolates is shown in Table 2. The distribution of antimicrobial resistance genes among the seven Salmonella enterica serotypes is shown schematically in Figure 2.
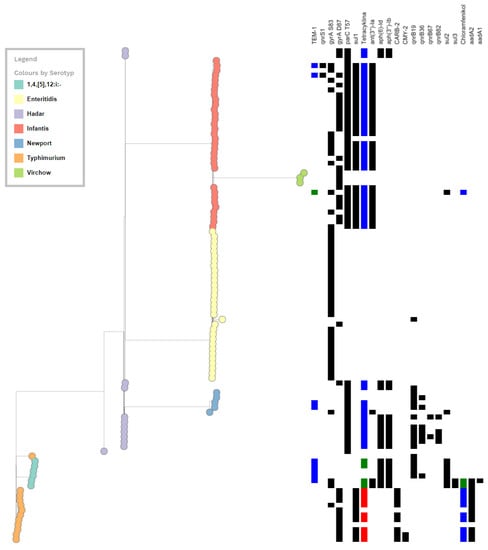
Figure 2.
Schematic diagram of the distribution of antimicrobial resistance genes among the 7 Salmonella enterica serotypes.
A total of five genes encoding β-lactamases were identified among the tested S. enterica, with the most common blaTEM-1 and blaCARB-2 alleles among S. Typhimurium (Table 2 and Tables S1–S7). Moreover, two S. Typhimurium isolates carried blaCMY-2, while one isolate had both, blaCMY-2 and blaCARB-2 (Table S1).
Six genes encoding aminoglycoside resistance–aac(6′)-Iaa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, aadA1 and aadA2–were detected (Table 2). Each of the seven Salmonella serotypes tested had at least one aminoglycoside resistance gene (Tables S1–S7).
Three distinct tet genes encoding tetracycline resistance efflux pumps were identified in the FQ-resistant Salmonella isolates tested. The tetA was the most common gene detected in 11 of 12 S. Hadar, 29 of 30 S. Infantis and 3 of 5 S. Newport. Additionally, 9 of 12 S. Typhimurium isolates and 4 of 6 monophasic 1,4,[5],12:i:- carried the tetG and tetB genes, respectively.
Sulphonamide resistance was predominantly encoded by three alleles–sul1, sul2 and sul3. The sul1 dominated in S. Typhimurium and S. Infantis, while sul2–in monophasic 1,4,[5],12:i:- S. Typhimurium (Figure 2, Tables S1–S7).
Phenicol resistance in the Salmonella isolates tested was encoded by two genes–floR and cmlA1 detected among 10 (9 S. Typhimurium and 1 S. Infantis) and two monophasic S. Typhimurium (Table 2 and Tables S1, S2, S4).
4. Discussion
There is an urgent need to preserve the effectiveness of antimicrobials for humans and animals, with the “One-health” initiative seeking to promote the responsible use of antimicrobials. Antimicrobial agents used in food-producing animals and in human medicine in Europe are frequently the same or belong to the same classes, and their use in both humans and animals might result in the selection of resistant clones, and the development of AMR [2]. Moreover, bacterial resistance to antimicrobials occurring in food-producing animals can spread to humans via food-borne routes, as has been observed for such zoonotic bacteria as Salmonella, by environmental contamination or through direct animal contact [2]. It is worth noting that, in accordance with Decision 2018/945/EU [21] on communicable diseases and related special health issues, antimicrobial resistance among Salmonella isolates in humans should be monitored. Furthermore, the WHO stated that the transmission of bacterial infections from non-human sources to humans, with disease-causing potential, is more evident in some bacteria (including non-typhoidal Salmonella) and the potential for such transmission should be recognised [5]. Nowadays, it is a well-recognised and generally acknowledged issue that fluoroquinolones are extensively used in medicine, veterinary and animal production [22,23].
In this study, we described the prevalence of FQ resistance among seven Salmonella enterica serotypes commonly occurring in humans in Poland. The 447 clinical human Salmonella enterica from different regions of Poland isolated in the years 2018–2019 were screened for FQ resistance. Our results indicated that there was a high level (37.6%) of FQ resistance among the isolates tested. As shown in Table 1, FQ resistance was most frequently detected among S. Hadar (92.3%), S. Virchow (75%) and S. Newport (71.4%). This was followed by S. Infantis (55.2%), S. Enteritidis (44.1%), monophasic 1,4,[5],12:i:- (16.2%) or S. Typhimurium (14.3%). Their ciprofloxacin MICs ranged from 0.125 to 3 mg/L. In accordance with the EFSA and ECDC joint report on antimicrobial resistance, in total 13.5% of Salmonella spp. isolated from humans were ciprofloxacin-resistant in Europe. Among them were: 20.9% Enteritidis, 19.8% Infantis, 5.6% monophasic Typhimurium and 5.1% Typhimurium, respectively [2].
Typically, chromosomal mutations inside the QRDRs of genes encoding DNA gyrase and topoisomerase IV, mainly the GyrA and/or ParC subunits, or the accumulation of mutations in these subunits, were considered to be important for ciprofloxacin resistance [8,11]. Out of the 100 Salmonella isolates resistant to ciprofloxacin (MIC > 0.06 mg/L) analysed by means of WGS in this study, 80 isolates had mutations in the gyrA QRDR, including 45 at Ser-83 and 35 at Asp-87. Both mutations, in GyrA and ParC, have been found among 34 Salmonella isolates tested. Notably, 30 S. Infantis with ciprofloxacin MIC ≥0.25 mg/L had an amino acid substitution in GyrA at codon 83 Ser→Tyr (n = 12) or codon 87 Asp→Tyr (n = 18) and a simultaneous alteration of ParC at codon 57 (Thr→Ser). Moreover, mutation Thr57→Ser in ParC was the only substitution detected among 47 tested Salmonella strains with ciprofloxacin MIC range 0.125–0.25 mg/L in S. Hadar, (0.19–0.75 mg/L) in S. Newport and (0.25–3 mg/L) in S. Infantis, respectively. This is in line with the reports of other authors that found Thr57Ser as the most prevalent mutation in ParC in NTS human isolates [14,24,25]. In accordance with a recent study, the ParC mutation at Thr57→Ser was detected as the main substitution in Salmonella enterica isolates from food-producing animals in China with a broad range of the ciprofloxacin MIC value (0.008–64 mg/L) [26]. Although authors suggest that the impact of the Thr57Ser mutation alone on quinolone susceptibility must be low, it may affect quinolone susceptibility when combined with the GyrA substitutions [26].
The plasmid-mediated quinolone resistance (PMQR) determinants are considered the cause of decreased susceptibility to FQs and factors facilitating the selection and acquisition of high-level FQ resistance [27]. In this study, qnr were the most prevalent PMQR genes detected among 7 of 12 S. Hadar, 4 of 5 S. Newport and 4 of 6 monophasic S. Typhimurium, 2 of 30 S. Infantis, 1 of 12 S. Typhimurium and 1 of 32 S. Enteritidis tested with ciprofloxacin MIC ≥0.19 mg/L, respectively. qnrB (variant: qnrB19, qnrB36, qnrB67 and qnrB82) was found in 17 isolates belonging to the serotypes Typhimurium (1/12), monophasic Typhimurium (4/6), Hadar (7/12), Newport (4/5), Enteritidis (1/32), while qnrS1 was found in 2 of 30 isolates of Infantis. Several studies have globally reported an association between the PMQR genes and the ciprofloxacin non-susceptible NTS isolates [14,25,28,29,30,31]. Additionally, the QRDR mutations and PMQR genes in NTS human isolates can be country specific. Similar to other reports, we detected NTS, including four monophasic Typhimurium, one Typhimurium and Enteritidis isolates, harbouring only PMQR genes without mutations in QRDR [14]. Interestingly, we identified three NTS strains (Hadar, Newport, monophasic Typhimurium 1,4,[5],12:i:-), three Hadar strains and one Newport harbouring two, three and four different variants of qnrB, respectively.
In addition to the variety of existing FQ resistance mechanisms, we found a high diversity of resistance genes by means of the in silico analysis of Salmonella WGS. Resistance to aminoglycosides was most common and showed the highest diversity (n = 6), including the aminoglycoside N-acetylotransferases, O-nucleotydylotransferases and O-phosphotransferases groups that modify aminoglycosides. In our study, aac(6′)-Iaa was the most frequently identified gene, in 68% (68/100) Salmonella isolates analysed by means of WGS. Less commonly found genes were: ant(3″)-Ia (n = 29) aph(3″)-Ib (n = 19) and aph(6)-Id (n = 18). The aadA1 and aadA2 genes also detected in our study confer aminoglycoside resistance, more specifically streptomycin and spectinomycin resistance. These resistance genes can be carried on mobile genetic elements that facilitate transfer between bacteria and promote dissemination within bacterial populations [32]. This finding corroborates several reports demonstrating the presence of multiple aminoglycoside resistance genes in Salmonella serovars [33,34]. Moreover, the “One-health” results from Canadian researchers [34] suggested that the use of lincomycin-spectinomycin in chickens may be selecting for gentamicin-resistant Salmonella in broilers, and then these resistant strains may be acquired by humans.
The resistance genes tet and sul that may confer resistance against tetracycline and sulphonamide, respectively, were also highly frequent among the WGS analysed Salmonella isolates, with the frequency amounting to 56% and 49%, respectively. Among them, tetA (n = 43) and sul1 (n = 39) were the most common. Based on the EFSA and ECDC joint report on antimicrobial resistance, in total 29% and 25.6% of Salmonella spp. isolated from humans in Europe are resistant to sulphonamides and tetracyclines, respectively [2]. The report also points to the high-level sulphamethoxazole resistance noted in broilers and turkeys (33.9% and 13.7%, respectively), and high-level tetracycline resistance (35.5% in broilers and 57.3% in turkeys, respectively) [2]. This may indicate that the indiscriminate use of antimicrobials in animal husbandry is also causing an increase in the antimicrobial resistance in Salmonella isolated from humans [35].
According to the ECDC and EFSA joint report [2], in 2019 in Europe, a high (25.8%) percentage of human Salmonella isolates were resistant to ampicillin, including monophasic Typhimurium (87.1%), Typhimurium (54.3%), Infantis (18.3%) and Enteritidis (8.1%). In our study, the blaTEM-1 gene encoding ampicillin resistance was detected in twelve (12%) Salmonella isolates belonging to monophasic 1,4,[5],12:i:- (n = 5), Infantis (n = 3), Newport (n = 2) and Typhimurium (n = 2). Interestingly, 8 isolates of 12 S. Typhimurium had the blaCARB-2 gene conferring resistance to ampicillin. The occurrence of the blaCARB-2 gene, earlier identified as blaPSE-1, in Salmonella has been limited to a few reports that concerned isolates from humans, animals and animal-derived products [36,37,38]. It is important to note that two S. Typhimurium isolates additionally carried blaCMY-2 (the acquired AmpC β-lactamase gene) that mediates resistance to a wide variety of beta-lactams, particularly cephalosporins. Our result is in line with the aforementioned ECDC and EFSA joint report where AmpC was identified in 0.1% of all the human Salmonella isolates tested [2].
5. Conclusions
In conclusion, our study demonstrated that a significant percentage of human Salmonella isolates is resistant to FQs. Although a small number of Salmonella strains was tested on the national scale, the results obtained illustrate the FQ resistance situation in Poland. The development of nontyphoidal Salmonella isolates resistant to FQs is of clinical importance since it may be associated with reduced effectiveness of therapies and represents a substantial public health concern. Zoonotic Salmonella is the leading foodborne pathogen, with global travel and food import significantly increasing the risk of acquiring salmonellosis. Moreover, our study strongly supports the need to carry out the surveillance and monitoring of AMR, including FQ resistance among human Salmonella isolates. In addition, in accordance with the “One Health” concept, collaboration and integrated surveillance for zoonotic Salmonella, including isolates originating from humans and food-producing animals, is necessary to elucidate trends in antimicrobial resistance. Additionally, it is worth remembering the role of the rational and prudent use of antibiotics including FQs, both in humans and animals. Because, as the Australian case indicated, banning the use of FQs in food-producing animals correlated with reduced FQ resistance in bacteria isolated from food, animals and patients [39].
Supplementary Materials
The following supporting information is downloaded at: https://www.mdpi.com/article/10.3390/pathogens12020193/s1, Table S1. The results of ciprofloxacin minimum inhibitory concentration, MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Typhimurium. Table S2. The results of ciprofloxacin minimum inhibitory concentration, MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant monophasic S. Typhimurium 1,4,[5],12:i:-. Table S3. The results of ciprofloxacin minimum inhibitory concentration (MIC), MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Hadar. Table S4. The results of ciprofloxacin minimum inhibitory concentration (MIC), MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Infantis. Table S5. The results of ciprofloxacin minimum inhibitory concentration (MIC), MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Newport. Table S6. The results of ciprofloxacin minimum inhibitory concentration (MIC), MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Virchow. Table S7. The results of ciprofloxacin minimum inhibitory concentration (MIC), MLST type and resistance genotype by means of WGS for clinical ciprofloxacin-resistant S. Enteritidis.
Author Contributions
K.P. was responsible for concept, design of this study and methods chosen, data analysis, check and manuscript draft. T.W. was responsible for performing WGS experiments, data analysis and participation in writing of WGS part of the manuscript. K.Z. was responsible for phenotypic analysis, including strain identification and serotyping, antibiotic susceptibility testing and performed WGS experiments. A.S. was responsible for performing WGS experiments. R.G. was the main reviewer and editor final version of manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by internal grants from the National Institute of Public Health–National Institute of Hygiene (no. 5/EM.1-2018; BB-1/2019; BB-1/2020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank M. Nowakowska and N. Wolaniuk for technical support involving antimicrobial susceptibility testing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella, Pasteur Institute: Paris, France, 2007. [Google Scholar]
- Authority, E.F.S. European Centre for Disease Prevention and Control the European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Burke, L.; Hopkins, K.L.; Meunier, D.; de Pinna, E.; Fitzgerald-Hughes, D.; Humphreys, H.; Woodford, N. Resistance to Third-Generation Cephalosporins in Human Non-Typhoidal Salmonella enterica Isolates from England and Wales, 2010–12. J. Antimicrob. Chemother. 2014, 69, 977–981. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-151552-8. [Google Scholar]
- Medicine, C.; For, V. Extralabel Use and Antimicrobials; FDA: Silver Spring, MD, USA, 2022.
- Van, T.T.H.; Nguyen, H.N.K.; Smooker, P.M.; Coloe, P.J. The Antibiotic Resistance Characteristics of Non-Typhoidal Salmonella Enterica Isolated from Food-Producing Animals, Retail Meat and Humans in South East Asia. Int. J. Food Microbiol. 2012, 154, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Yanat, B.; Rodríguez-Martínez, J.-M.; Touati, A. Plasmid-Mediated Quinolone Resistance in Enterobacteriaceae: A Systematic Review with a Focus on Mediterranean Countries. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2017, 36, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Xu, X.; Gu, Z.; Meng, J.; Wufuer, X.; Wang, M.; Huang, M.; Chen, J.; Jing, C.; Xiong, Z.; et al. Molecular Epidemiology and Antimicrobial Resistance of Invasive Non-Typhoidal Salmonella in China, 2007–2016. Infect. Drug Resist. 2019, 12, 2885–2897. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Hooper, D.C. Mechanisms of Fluoroquinolone Resistance. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 1999, 2, 38–55. [Google Scholar] [CrossRef]
- Piekarska, K.; Wołkowicz, T.; Zacharczuk, K.; Rzeczkowska, M.; Chróst, A.; Bareja, E.; Olak, M.; Gierczyński, R. Co-Existence of Plasmid-Mediated Quinolone Resistance Determinants and Mutations in GyrA and ParC among Fluoroquinolone-Resistant Clinical Enterobacteriaceae Isolated in a Tertiary Hospital in Warsaw, Poland. Int. J. Antimicrob. Agents 2015, 45, 238–243. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-Mediated Quinolone Resistance. Microbiol. Spectr. 2014, 2, 475–503. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.-B.; Lauderdale, T.-L.Y.; Huang, C.-H.; Chang, P.-R.; Wang, Y.-H.; Shigemura, K.; Lin, Y.-H.; Chang, W.-C.; Wang, K.-C.; Huang, T.-W.; et al. Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan. Antibiotics 2021, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Salmonellosis-Annual Epidemiological Report for 2017. Available online: https://www.ecdc.europa.eu/en/publications-data/salmonellosis-annual-epidemiological-report-2017 (accessed on 24 October 2022).
- Wołkowicz, T.; Zacharczuk, K.; Gierczyński, R.; Nowakowska, M.; Piekarska, K. Antimicrobial Resistance and Whole-Genome Characterisation of High-Level Ciprofloxacin-Resistant Salmonella Enterica Serovar Kentucky ST 198 Strains Isolated from Human in Poland. Int. J. Mol. Sci. 2021, 22, 9381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Deatherage Kaiser, B.L.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
- Official Journal of the European Union. Commission Implementing Decision (EU) 2018/945 of 22 June 2018 on the Communicable Diseases and Related Special Health Issues to Be Covered by Epidemiological Surveillance as Well as Relevant Case Definitions (Text with EEA Relevance); Official Journal of the European Union: Luxembourg, 2018; Volume 170. [Google Scholar]
- Apreja, M.; Sharma, A.; Balda, S.; Kataria, K.; Capalash, N.; Sharma, P. Antibiotic Residues in Environment: Antimicrobial Resistance Development, Ecological Risks, and Bioremediation. Environ. Sci. Pollut. Res. 2022, 29, 3355–3371. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Yassin, H.; Fels-Klerx, H.J.; Berendsen, B.J.A. RIKILT-BU Toxicology Bioassays & Novel Foods, VLAG, RIKILT-Business unit Dierbehandelingsmiddelen. Antibiotic Residues and Resistance in the Environment; Pikkemaat, M.G., Yassin, H., Fels-Klerx, H.J., Berendsen, B.J.A., Eds.; RIKILT Wageningen UR: Wageningen, The Netherlands, 2016. [Google Scholar]
- Campos, M.J.; Palomo, G.; Hormeño, L.; Herrera-León, S.; Domínguez, L.; Vadillo, S.; Píriz, S.; Quesada, A. Prevalence of Quinolone Resistance Determinants in Non-Typhoidal Salmonella Isolates from Human Origin in Extremadura, Spain. Diagn. Microbiol. Infect. Dis. 2014, 79, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Gunell, M.; Webber, M.A.; Kotilainen, P.; Lilly, A.J.; Caddick, J.M.; Jalava, J.; Huovinen, P.; Siitonen, A.; Hakanen, A.J.; Piddock, L.J.V. Mechanisms of Resistance in Nontyphoidal Salmonella Enterica Strains Exhibiting a Nonclassical Quinolone Resistance Phenotype. Antimicrob. Agents Chemother. 2009, 53, 3832–3836. [Google Scholar] [CrossRef]
- Chang, M.-X.; Zhang, J.-F.; Sun, Y.-H.; Li, R.-S.; Lin, X.-L.; Yang, L.; Webber, M.A.; Jiang, H.-X. Contribution of Different Mechanisms to Ciprofloxacin Resistance in Salmonella Spp. Front. Microbiol. 2021, 12, 663731. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-Mediated Quinolone Resistance: Two Decades On. Drug Resist. Updat. 2016, 29, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Acheampong, G.; Owusu, M.; Owusu-Ofori, A.; Osei, I.; Sarpong, N.; Sylverken, A.; Kung, H.-J.; Cho, S.-T.; Kuo, C.-H.; Park, S.E.; et al. Chromosomal and Plasmid-Mediated Fluoroquinolone Resistance in Human Salmonella Enterica Infection in Ghana. BMC Infect. Dis. 2019, 19, 898. [Google Scholar] [CrossRef]
- Karp, B.E.; Campbell, D.; Chen, J.C.; Folster, J.P.; Friedman, C.R. Plasmid-Mediated Quinolone Resistance in Human Nontyphoidal Salmonella Infections: An Emerging Public Health Problem in the United States. Zoonoses Public Health 2018, 65, 838–849. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Lee, S.-K.; Park, M.-S.; Na, H.-T. Analysis of the Fluoroquinolone Antibiotic Resistance Mechanism of Salmonella Enterica Isolates. J. Microbiol. Biotechnol. 2016, 26, 1605–1612. [Google Scholar] [CrossRef]
- Lee, S.; Park, N.; Yun, S.; Hur, E.; Song, J.; Lee, H.; Kim, Y.; Ryu, S. Presence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Non-Typhoidal Salmonella Strains with Reduced Susceptibility to Fluoroquinolones Isolated from Human Salmonellosis in Gyeonggi-Do, South Korea from 2016 to 2019. Gut Pathog. 2021, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. MedChemComm 2016, 7, 11–27. [Google Scholar] [CrossRef]
- Rodrigues, G.L.; Panzenhagen, P.; Ferrari, R.G.; dos Santos, A.; Paschoalin, V.M.F.; Conte-Junior, C.A. Frequency of Antimicrobial Resistance Genes in Salmonella from Brazil by in Silico Whole-Genome Sequencing Analysis: An Overview of the Last Four Decades. Front. Microbiol. 2020, 11, 1864. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.W.; Parmley, E.J.; Avery, B.P.; Irwin, R.J.; Reid-Smith, R.J.; Deckert, A.E.; Finley, R.L.; Daignault, D.; Alexander, D.C.; Allen, V.; et al. A One-Health Genomic Investigation of Gentamicin Resistance in Salmonella from Human and Chicken Sources in Canada, 2014 to 2017. Antimicrob. Agents Chemother. 2021, 65, e00966. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; Habib, I.; Mezal, E.H.; Kareem, H.S.; Laird, T.; O’Dea, M.; Abraham, S. Occurrence, Antimicrobial Resistance and Whole-Genome Sequencing Analysis of Salmonella Isolates from Chicken Carcasses Imported into Iraq from Four Different Countries. Int. J. Food Microbiol. 2018, 284, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Randall, L.P.; Cooles, S.W.; Osborn, M.K.; Piddock, L.J.V.; Woodward, M.J. Antibiotic Resistance Genes, Integrons and Multiple Antibiotic Resistance in Thirty-Five Serotypes of Salmonella Enterica Isolated from Humans and Animals in the UK. J. Antimicrob. Chemother. 2004, 53, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Llanes, C.; Kirchgesner, V.; Plesiat, P. Propagation of TEM- and PSE-Type β-Lactamases among Amoxicillin-Resistant Salmonella Spp. Isolated in France. Antimicrob. Agents Chemother. 1999, 43, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.C.; Turnidge, J.; Collignon, P.; Looke, D.; Barton, M.; Gottlieb, T. Control of Fluoroquinolone Resistance through Successful Regulation, Australia. Emerg. Infect. Dis. 2012, 18, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
