Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families
Abstract
1. Introduction
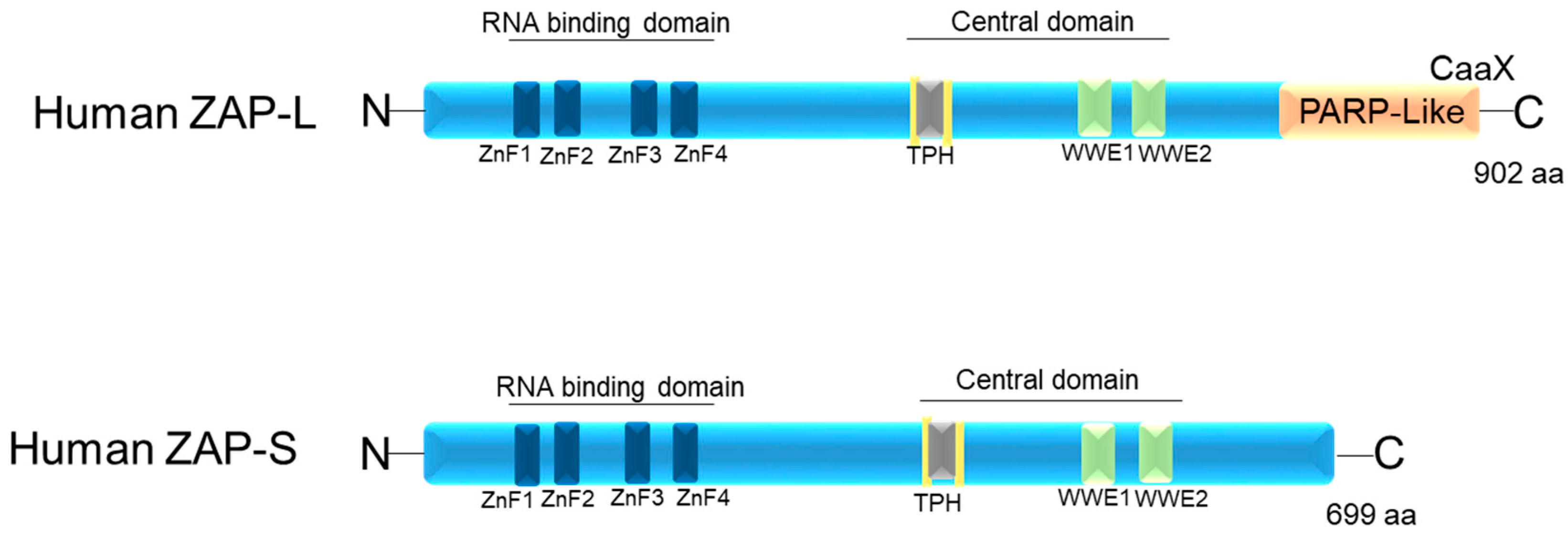
2. Zinc Finger Antiviral Protein—ZAP
3. RNA Recognition by ZAP
4. Cofactors Required by ZAP for its Antiviral Activity
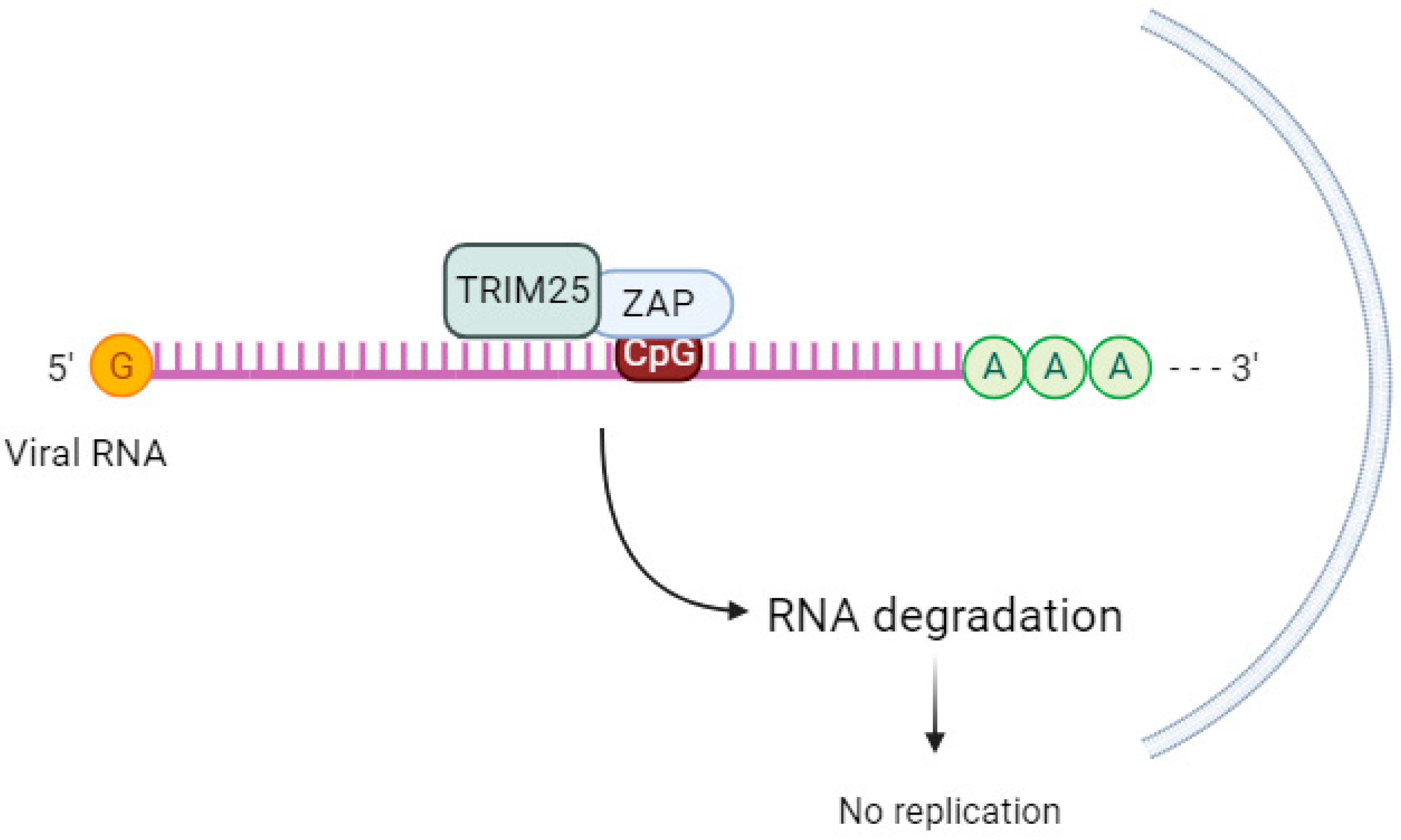
4.1. Tripartite Motif Containing 25 (TRIM25)
4.2. KH and NYN Domain Containing (KHNYN)
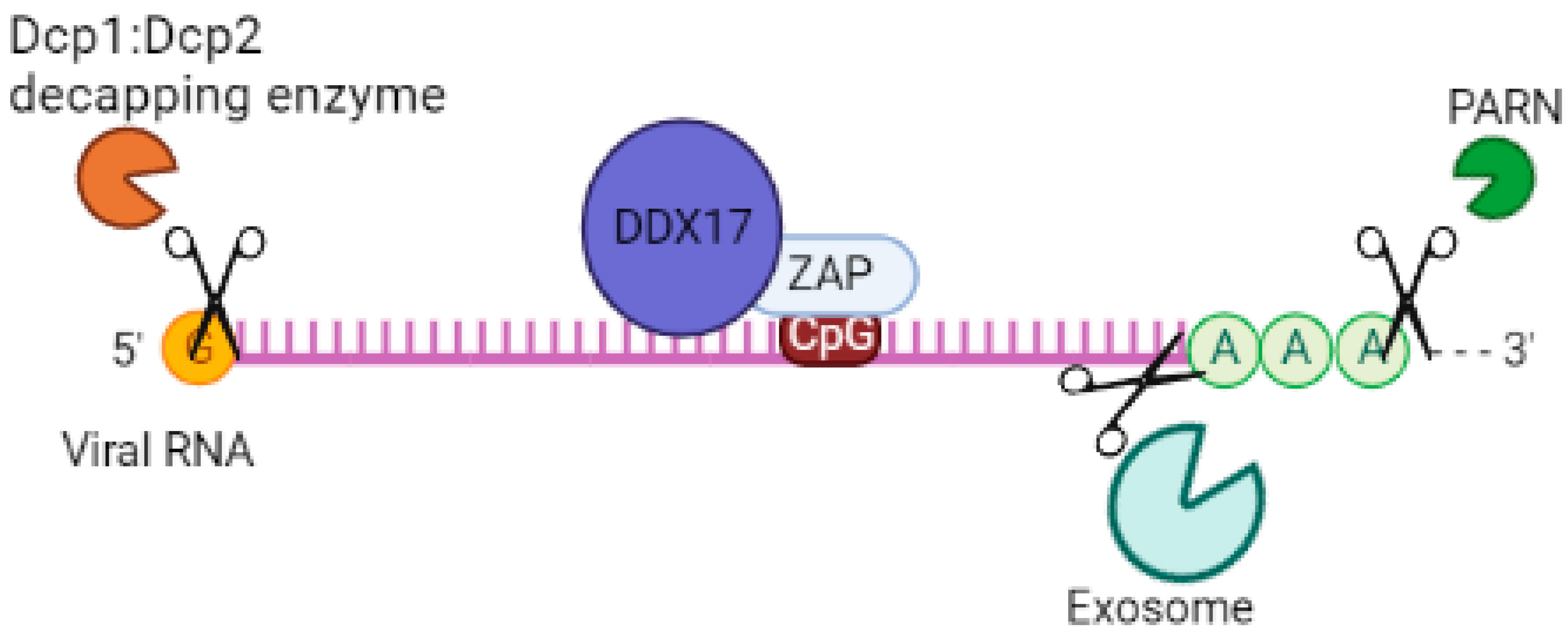
4.3. Exosome
4.4. p72 RNA Helicase
5. ZAP Protein Regulators
5.1. Matrin 3 (MATR3)
5.2. Glycogen Synthase Kinase 3β (GSK3β)
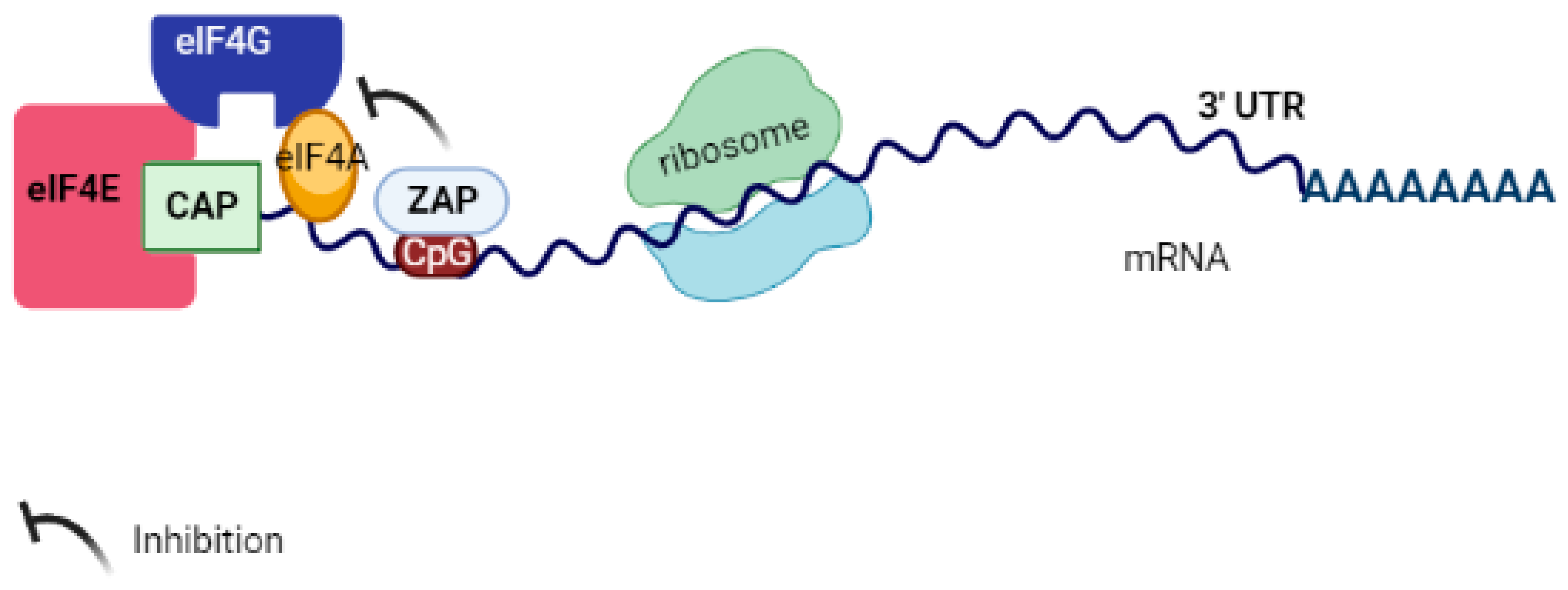
6. ZAP Inhibits Target Virus RNA Translation
7. Immune Pathways Associated with Antiviral Activity of ZAP
7.1. Type I and III Interferons (IFNs)
7.2. Retinoic Acid-Inducible Gene I (RIG-I)
7.3. OAS1–Rnasel Antiviral Pathway
8. Antiviral Activity of ZAP in Different Virus Families
8.1. Retroviridae Family
8.1.1. Moloney and Murine Leukemia Virus (MoMLV or MuLV or MLV)
8.1.2. Human Immunodeficiency Virus Type 1 (HIV-1)
8.1.3. Avian Leukosis Virus Subgroup J (ALV-J)
8.1.4. Human T-lymphotropic Virus Type 1 (HTLV-1)
8.2. Flaviviridae Family
8.2.1. Japanese Encephalitis Virus (JEV)
8.2.2. Zika Virus (ZIKV)
8.3. Togaviridae Family
8.3.1. Sindbis Virus (SINV)
8.3.2. Semliki Forest Virus (SFV)
8.4. Filoviridae Family
Ebola Virus (EBOV) and Marburg Virus (MARV)
8.5. Picornaviridae Family
8.5.1. Coxsackievirus B3
8.5.2. Echovirus 7 (E7)
8.5.3. Enterovirus A71 (EV-A71)
8.5.4. Poliovirus
8.6. Orthomyxoviridae Family
Influenza A Virus
8.7. Poxviridae Family
Vaccinia Virus Ankara (MVA)
8.8. Hepadnaviridae Family
Hepatitis B Virus
8.9. Herpesviridae Family
Murine Gammaherpesvirus 68
8.10. Coronaviridae Family
8.11. Paramyxoviridae Family
8.11.1. Sendai Virus
8.11.2. Newcastle Disease Virus (NDV)
9. Challenges and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chow, K.T.; Gale, M.; Loo, Y.-M. RIG-I and Other RNA Sensors in Antiviral Immunity. Annu. Rev. Immunol. 2018, 36, 667–694. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, E.; Glaunsinger, B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 2015, 479-480, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, F.; Zhu, Y.; Gao, G. Zinc-finger antiviral protein inhibits XMRV infection. PLoS ONE 2012, 7, 39159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muller, S.; Möller, P.; Bick, M.J.; Wurr, S.; Becker, S.; Günther, S.; Kümmerer, B.M. Inhibition of filovirus replication by the zinc finger antiviral protein. Virol. J. 2007, 81, 2391–2400. [Google Scholar] [CrossRef] [PubMed]
- Bick, M.J.; Carroll, J.-W.N.; Gao, G.; Goff, S.P.; Rice, C.M.; MacDonald, M.R. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. Virol. J. 2003, 77, 11555–11562. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Goff, S.P.; Gao, G. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 2012, 31, 4236–4246. [Google Scholar] [CrossRef]
- Yang, E.; Nguyen, L.P.; Wisherop, C.A.; Kan, R.L.; Li, M.M.H. The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. Front. Cell. Infect. Microbiol. 2022, 12, 886929. [Google Scholar] [CrossRef]
- Chiu, H.-P.; Chiu, H.; Yang, C.-F.; Lee, Y.-L.; Chiu, F.-L.; Kuo, H.-C. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLOS Pathog. 2018, 14, 1007166. [Google Scholar] [CrossRef]
- Wang, N.; Dong, Q.; Li, J.; Jangra, R.K.; Fan, M.; Brasier, A.R.; Lemon, S.M.; Pfeffer, L.M.; Li, K. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. J. Biol. Chem. 2010, 285, 6080–6090. [Google Scholar] [CrossRef] [PubMed]
- Galão, R.P.; Wilson, H.; Schierhorn, K.L.; Debeljak, F.; Bodmer, B.S.; Goldhill, D.; Hoenen, T.; Wilson, S.J.; Swanson, C.M.; Neil, S.J.D. TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction. PLoS Pathog. 2022, 18, e1010530. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Guo, X.; Goff, S.P. Inhibition of Retroviral RNA Production by ZAP, a CCCH-Type Zinc Finger Protein. Science 2002, 297, 1703–1706. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yan, K.; Wei, L.; Yang, J.; Lu, C.; Xiong, F.; Zheng, C.; Xu, W. Zinc finger antiviral protein inhibits coxsackievirus B3 virus replication and protects against viral myocarditis. Antivir. Res. 2015, 123, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Kerns, J.A.; Emerman, M.; Malik, H.S. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008, 4, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.H.; Aguilar, E.G.; Michailidis, E.; Pabon, J.; Park, P.; Wu, X.; de Jong, Y.P.; Schneider, W.M.; Molina, H.; Rice, C.M.; et al. Characterization of novel splice variants of zinc finger antiviral protein (ZAP). J. Virol. 2019, 93, e00715-19. [Google Scholar] [CrossRef] [PubMed]
- Schwerk, J.; Soveg, F.W.; Ryan, A.P.; Thomas, K.R.; Hatfield, L.D.; Ozarkar, S.; Forero, A.; Kell, A.M.; Roby, J.A.; So, L.; et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 2019, 20, 1610–1620. [Google Scholar] [CrossRef]
- Charron, G.; Li, M.M.; MacDonald, M.R.; Hang, H.C. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc. Natl. Acad. Sci. USA 2013, 110, 11085–11090. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Gao, Y.; Zhu, J.; Liu, S.; Gao, G.; Gao, P. Molecular Mechanism of RNA Recognition by ZincFinger Antiviral Protein. Cell Rep. 2020, 30, 46–52.e4. [Google Scholar] [CrossRef]
- Li, M.M.H.; Lau, Z.; Cheung, P.; Aguilar, E.G.; Schneider, W.M.; Bozzacco, L.; Molina, H.; Buehler, E.; Takaoka, A.; Rice, C.M.; et al. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLoS Pathog. 2017, 13, 1006145. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Tu, F.; Wang, Q.; Fan, Z.; Gao, G. TRIM25 Is Required for the Antiviral Activity of Zinc Finger Antiviral Protein. Diamond MS, editor. J. Virol. 2017, 91, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defense targeting non-self RNA. Nature 2017, 550, 7674. [Google Scholar] [CrossRef] [PubMed]
- Colmant, A.M.G.; Hobson-Peters, J.; Slijkerman, T.A.P.; Harrison, J.J.; Pijlman, G.P.; Van Oers, M.M.; Simmonds, P.; Hall, R.A.; Fros, J.J. Insect-Specific Flavivirus Replication in Mammalian Cells Is Inhibited by Physiological Temperature and the Zinc-Finger Antiviral Protein. Viruses 2021, 13, 573. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M.M.; Kibe, A.; Rand, U.; Pekarek, L.; Ye, L.; Buck, S.; Smyth, R.P.; Cicin-Sain, L.; Caliskan, N. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat. Commun. 2021, 12, 7193. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Lista, M.J.; Ficarelli, M.; Swanson, C.M.; Neil, S.J.D. S-farnesylation is essential for antiviral activity of the long ZAP isoform against RNA viruses with diverse replication strategies. PLoS Pathog. 2021, 17, e1009726. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Visser, I.; Tang, B.; Yan, K.; Nakayama, E.; Visser, T.M.; Koenraadt, C.J.M.; van Oers, M.M.; Pijlman, G.P.; Suhrbier, A.; et al. The dinucleotide composition of the Zika virus genome is shaped by conflicting evolutionary pressures in mammalian hosts and mosquito vectors. PLoS Biol. 2021, 19, e3001201. [Google Scholar] [CrossRef]
- Odon, V.; Fros, J.J.; Goonawardane, N.; Dietrich, I.; Ibrahim, A.; Alshaikhahmed, K.; Nguyen, D.; Simmonds, P. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res. 2019, 47, 8061–8083. [Google Scholar] [CrossRef]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P.A. Systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013, 4, 2240. [Google Scholar] [CrossRef]
- Karlberg, T.; Klepsch, M.; Thorsell, A.G.; Andersson, C.D.; Linusson, A.; Schuler, H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J. Biol. Chem. 2015, 290, 7336–7344. [Google Scholar] [CrossRef]
- Guo, X.; Carroll, J.-W.N.; MacDonald, M.R.; Goff, S.P.; Gao, G. The Zinc Finger Antiviral Protein Directly Binds to Specific Viral mRNAs through the CCCH Zinc Finger Motifs. J. Virol. 2004, 78, 12781–12787. [Google Scholar] [CrossRef]
- Mao, R.; Nie, H.; Cai, D.; Zhang, J.; Liu, H.; Yan, R.; Cuconati, A.; Block, T.M.; Guo, J.-T.; Guo, H. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 2013, 9, e1003494. [Google Scholar] [CrossRef] [PubMed]
- Todorova, T.; Bock, F.; Chang, P. PARP13 and RNA regulation in immunity and cancer. Trends Mol. Med. 2015, 21, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.; Liu, L.; Shen, S.; Deng, H.; Gao, G. Zinc finger antiviral protein inhibits murine gammaherpesvirus 68 M2 expression and regulates viral latency in cultured cells. Virol. J. 2012, 86, 12431–12434. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.E.; Karpala, A.J.; Ward, A.; Bean, A.G. Characterisation of chicken ZAP. Dev. Comp. Immunol. 2014, 46, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Carneiro, D.; Takata, M.A.; Ong, H.; Shilton, A.; Bieniasz, P.D. Origin and evolution of the zinc finger antiviral protein. PLoS Pathog. 2021, 17, e1009545. [Google Scholar] [CrossRef]
- Leung, A.K.; Vyas, S.; Rood, J.E.R.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007, 81, 11246–11255. [Google Scholar] [CrossRef]
- MacDonald, M.R.; Machlin, E.S.; Albin, O.R.; Levy, D.E. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 2007, 81, 13509–13518. [Google Scholar] [CrossRef]
- Ryman, K.D.; Meier, K.C.; Nangle, E.M.; Ragsdale, S.L.; Korneeva, N.L.; Rhoads, R.E.; MacDonald, M.R.; Klimstra, W.B. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J. Virol. 2005, 79, 1487–1499. [Google Scholar] [CrossRef]
- Hayakawa, S.; Shiratori, S.; Yamato, H.; Kameyama, T.; Kitatsuji, C.; Kashigi, F.; Goto, S.; Kameoka, S.; Fujikura, D.; Yamada, T.; et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 2011, 12, 37–44. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.-Y.; Krug, R.M.; Sullivan, C.S. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 2013, 14, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Todorova, T.; Bock, F.J.; Chang, P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 Transcript. Nat. Commun. 2014, 5, 5362. [Google Scholar] [CrossRef] [PubMed]
- Okudera, M.; Odawara, M.; Arakawa, M.; Kawaguchi, S.; Seya, K.; Matsumiya, T.; Sato, R.; Ding, J.; Morita, E.; Imaizumi, T. Expression of Zinc-Finger Antiviral Protein in hCMEC/D3 Human Cerebral Microvascular Endothelial Cells: Effect of a Toll-Like Receptor 3 Agonist. Neuroimmunomodulation 2022, 29, 349–358. [Google Scholar] [CrossRef]
- Zhu, Y.; Gao, G. ZAP-mediated mRNA degradation. RNA Biol. 2008, 5, 65–67. [Google Scholar] [CrossRef]
- Gonzalez-Perez, A.C.; Stempel, M.; Wyler, E.; Urban, C.; Piras, A.; Hennig, T.; Ganskih, S.; Wei, Y.; Heim, A.; Landthaler, M.; et al. The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral UL4/UL5 Transcripts. mBio 2021, 12, e02683-20. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L.; Pereira, G.C.; Cheung, L.E.; Rose, R.J.; Kazazian, H.H.J. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 2015, 11, e1005252. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Braczyk, K.; Gonçalves-Carneiro, D.; Dawidziak, D.M.; Sanchez, K.; Ong, H.; Wan, Y.; Zadrozny, K.K.; Ganser-Pornillos, B.K.; Bieniasz, P.D.; et al. Poly(ADP-ribose) potentiates ZAP antiviral activity. PLoS Pathog. 2022, 18, e1009202. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y.; Zhang, K.; Wang, X.; Sun, J.; Gao, G.; Liu, Y. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat. Struct. Mol. Biol. 2012, 19, 430–435. [Google Scholar] [CrossRef]
- Soveg, F.W.; Schwerk, J.; Gokhale, N.S.; Cerosaletti, K.; Smith, J.R.; Pairo-Castineira, E.; Kell, A.M.; Forero, A.; Zaver, S.A.; Esser-Nobis, K.; et al. Endomembrane targeting of human OAS1 p46 augments antiviral activity. eLife 2021, 10, e71047. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A Prenylated dsRNA Sensor Protects Against Severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef]
- Nchioua, R.; Kmiec, D.; Muller, J.A.; Conzelmann, C.; Gross, R.; Swanson, C.M.; Neil, S.J.D.; Stenger, S.; Sauter, D.; Münch, J.; et al. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio 2020, 11, e01930-20. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Nchioua, R.; Sherrill-Mix, S.; Sturzel, C.M.; Heusinger, E.; Braun, E.; Gondim, M.V.P.; Hotter, D.; Sparrer, K.M.J.; Hahn, B.H.; et al. CpG Frequency in the 5′ Third of the env Gene Determines Sensitivity of Primary HIV-1 Strains to the Zinc-Finger Antiviral Protein. mBio 2020, 11, e02903-19. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.M.J.; Albin, O.R.; Carroll, J.W.N.; Jones, C.T.; Rice, C.M.; MacDonald, M.R. Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. J. Virol. 2010, 84, 4504–4512. [Google Scholar]
- Meagher, J.L.; Takata, M.; Gonçalves-Carneiro, D.; Keane, S.C.; Rebendenne, A.; Ong, H.; Orr, V.K.; MacDonald, M.R.; Stuckey, J.A.; Bieniasz, P.D.; et al. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CGrich viral sequences. Proc. Natl. Acad. Sci. USA 2019, 116, 24303–24309. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, G.; Lv, F.; Wang, X.; Ji, X.; Xu, Y.; Sun, J.; Wu, L.; Zheng, Y.-T.; Gao, G. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 15834–15839. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N.; Gerber-Huber, S. DNA methylation and CpG suppression. Cell. Differ. 1985, 17, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, X.; Gao, G. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell 2010, 1, 752–759. [Google Scholar] [CrossRef]
- Nguyen, L.P.; Aldana, K.S.; Yang, E.; Yao, Z.; Li, M.M.H. Alphavirus Evasion of Zinc Finger Antiviral Protein (ZAP) Correlates with CpG Suppression in a Specific Viral nsP2 Gene Sequence. Viruses 2023, 15, 830. [Google Scholar] [CrossRef]
- Goonawardane, N.; Nguyen, D.; Simmonds, P. Association of Zinc Finger Antiviral Protein Binding to Viral Genomic RNA with Attenuation of Replication of Echovirus 7. mSphere 2021, 6, e01138-20. [Google Scholar] [CrossRef]
- Gonçalves-Carneiro, D.; Mastrocola, E.; Lei, X.; DaSilva, J.; Chan, Y.F.; Bieniasz, P.D. Rational attenuation of RNA viruses with zinc finger antiviral protein. Nat. Microbiol. 2022, 7, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. PNAS 2007, 104, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lou, D.I.; Sawyer, S.L. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011, 7, e1002388. [Google Scholar] [CrossRef] [PubMed]
- Martín-Vicente, M.; Medrano, L.M.; Resino, S.; García-Sastre, A.; Martínez, I. TRIM25 in the Regulation of the Antiviral Innate Immunity. Front. Immunol. 2017, 22, 1187. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Bergamaschi, A.; Luna, J.M.; David, A.; Uchil, P.D.; Margottin-Goguet, F.; Mothes, W.; Hazan, U.; Transy, C.; Pancino, G.; et al. Human TRIM gene expression in response to interferons. PLoS ONE 2009, 4, e4894. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.G.; Sparrer, K.M.J.; Chiang, C.; Reis, R.A.; Chiang, J.J.; Zurenski, M.A.; Wan, Y.; Gack, M.U.; Pornillos, O. TRIM25 Binds RNA to Modulate Cellular Anti-viral Defense. J. Mol. Biol. 2018, 430, 5280–5293. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017, 15, e2004086. [Google Scholar] [CrossRef]
- Yang, E.; Huang, S.; Jami-Alahmadi, Y.; McInerney, G.M.; Wohlschlegel, J.A.; Li, M.M.H. Elucidation of TRIM25 ubiquitination targets involved in diverse cellular and antiviral processes. PLoS Pathog. 2022, 18, e1010743. [Google Scholar] [CrossRef]
- Choudhury, N.R.; Heikel, G.; Trubitsyna, M.; Kubik, P.; Nowak, J.S.; Webb, S.; Granneman, S.; Spanos, C.; Rappsilber, J.; Castello, A.; et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 2017, 15, 10. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Chiweshe, S.; McCormick, D.; Raper, A.; Wickenhagen, A.; DeFillipis, V.; Gaunt, E.; Simmonds, P.; Wilson, S.J.; Grey, F. Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog. 2020, 16, e1008844. [Google Scholar] [CrossRef]
- Ficarelli, M.; Wilson, H.; Pedro Galão, R.; Mazzon, M.; Antzin-Anduetza, I.; Marsh, M.; Neil, S.J.; Swanson, C.M. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. eLife 2019, 8, e46767. [Google Scholar] [CrossRef] [PubMed]
- Malgras, M.; Garcia, M.; Jousselin, C.; Bodet, C.; Lévêque, N. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses 2021, 13, 582. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Jensen, T.H. The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci. 2008, 33, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Brodersen, D.E.; Jensen, T.H. Origins and activities of the eukaryotic exosome. J. Cell Sci. 2009, 122, 1487–1494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping: The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef]
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215. [Google Scholar] [CrossRef]
- Wortham, N.C.; Ahamed, E.; Nicol, S.M.; Thomas, R.S.; Periyasamy, M.; Jiang, J.; Ochocka, A.M.; Shousha, S.; Huson, L.; Bray, S.E.; et al. The DEAD-box protein p72 regulates ERa-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERa-positive breast cancer. Oncogene 2009, 28, 4053–4064. [Google Scholar] [CrossRef]
- Honig, A.; Auboeuf, D.; Parker, M.M.; O’Malley, B.W.; Berget, S.M. Regulation of alternative splicing by the ATP-dependent DEADbox RNA helicase p72. Mol. Cell. Biol. 2002, 22, 5698–5707. [Google Scholar] [CrossRef]
- Chen, G.; Guo, X.; Lv, F.; Xu, Y.; Gao, G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad. Sci. USA 2008, 105, 4352–4357. [Google Scholar] [CrossRef]
- Belgrader, P.; Dey, R.; Berezney, R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J. Biol. Chem. 1991, 266, 9893–9899. [Google Scholar] [CrossRef]
- Malik, A.M.; Miguez, R.A.; Li, X.; Ho, Y.-S.; Feldman, E.L.; Barmada, S.J. Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization. eLife 2018, 17, e35977. [Google Scholar] [CrossRef]
- Salton, M.; Elkon, R.; Borodina, T.; Davydov, A.; Yaspo, M.-L.; Halperin, E.; Shiloh, Y. Matrin 3 Binds and Stabilizes mRNA. PLoS ONE 2011, 6, e23882. [Google Scholar] [CrossRef]
- Erazo, A.; Goff, S.P. Nuclear matrix protein Matrin 3 is a regulator of ZAP-mediated retroviral restriction. Retrovirology 2015, 12, 57. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Ostheimer, G.J.; van Vugt, M.A.; Jørgensen, C.; Miron, I.M.; Diella, F.; Colwill, K.; Taylor, L.; Elder, K.; et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007, 129, 1415–1426. [Google Scholar] [CrossRef]
- Sun, L.; Lv, F.X.; Guo, X.M.; Gao, G.X. Glycogen Synthase Kinase 3 beta (GSK3 beta) Modulates Antiviral Activity of Zincfinger Antiviral Protein (ZAP). J. Biol. Chem. 2012, 287, 22882–22888. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Witteveldt, J.; Evans, D.J.; Simmonds, P. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic Acids Res. 2014, 42, 4527–4545. [Google Scholar] [CrossRef]
- Lozhkov, A.A.; Klotchenko, S.A.; Ramsay, E.S.; Moshkoff, H.D.; Moshkoff, D.A.; Vasin, A.V.; Salvato, M.S. The Key Roles of Interferon Lambda in Human Molecular Defense against Respiratory Viral Infections. Pathogens 2020, 9, 989. [Google Scholar] [CrossRef]
- Rouse, B.T.; Sehrawat, S. Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 2010, 10, 514–526. [Google Scholar] [CrossRef]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef]
- Crosse, K.M.; Monson, E.A.; Beard, M.R.; Helbig, K.J. Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun. 2018, 10, 85–93. [Google Scholar] [CrossRef]
- Yang, E.; Li, M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere with Viral RNA Processes. Front. Immunol. 2020, 11, 605024. [Google Scholar] [CrossRef]
- Gregersen, L.H.; Mitter, R.; Ugalde, A.P.; Nojima, T.; Proudfoot, N.J.; Agami, R.; Stewart, A.; Svejstrup, J.Q. SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell 2019, 177, 1797–1813.e18. [Google Scholar] [CrossRef]
- Glasker, S.; Toller, M.; Kummerer, B.M. The alternate triad motif of the poly(ADPribose) polymerase-like domain of the human zinc finger antiviral protein is essential for its antiviral activity. J. Gen. Virol. 2014, 95, 816–822. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Oshiumi, H.; Miyashita, M.; Matsumoto, M.; Seya, T. A Distinct Role of Riplet-Mediated K63-Linked Polyubiquitination of the RIG-I Repressor Domain in Human Antiviral Innate Immune Responses. PLoS Pathog. 2013, 9, e1003533. [Google Scholar] [CrossRef]
- Cadena, C.; Ahmad, S.; Xavier, A.; Willemsen, J.; Park, S.; Park, J.W.; Oh, S.W.; Fujita, T.; Hou, F.; Binder, M.; et al. Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 2019, 177, 1187–1200.e16. [Google Scholar] [CrossRef]
- Buckmaster, M.V.; Goff, S.P. Riplet Binds the Zinc Finger Antiviral Protein (ZAP) and Augments ZAP-Mediated Restriction of HIV-1. J. Virol. 2022, 96, e0052622. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Lee, H.; Komano, J.; Saitoh, Y.; Yamaoka, S.; Kozaki, T.; Misawa, T.; Takahama, M.; Satoh, T.; Takeuchi, O.; Yamamoto, N.; et al. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc. Natl. Acad. Sci. USA 2013, 110, 12379–12384. [Google Scholar] [CrossRef]
- Lian, H.; Zang, R.; Wei, J.; Ye, W.; Hu, M.-M.; Chen, Y.-D.; Zhang, X.-N.; Guo, Y.; Lei, C.-Q.; Yang, Q.; et al. The Zinc-Finger Protein ZCCHC3 Binds RNA and Facilitates Viral RNA Sensing and Activation of the RIG-I-like Receptors. Immunity 2018, 49, 438–448.e5. [Google Scholar] [CrossRef]
- Schwartz, S.L.; Park, E.N.; Vachon, V.K.; Danzy, S.; Lowen, A.C.; Conn, G.L. Human OAS1 activation is highly dependent on both RNA sequence and context of activating RNA motifs. Nucleic Acids Res. 2020, 48, 7520–7531. [Google Scholar] [CrossRef]
- Bhattacharyya, S. Can’t RIDD off viruses. Front. Microbiol. 2014, 5, 292. [Google Scholar] [CrossRef]
- Silverman, R.H.; Weiss, S.R. Viral phosphodiesterases that antagonize doublestranded RNA signaling to RNase L by degrading 2–5A. J. Interferon Cytokine Res. 2014, 34, 455–463. [Google Scholar] [CrossRef]
- Karki, S.; Li, M.M.H.; Schoggins, J.W.; Tian, S.; Rice, C.M.; MacDonald, M.R. Multiple Interferon Stimulated Genes Synergize with the Zinc Finger Antiviral Protein to Mediate Anti-Alphavirus Activity. PLoS ONE 2012, 7, e37398. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.M.H.; Zhao, J.; Li, S.; MacDonald, M.R.; Rice, C.M.; Gao, X.; Gao, G. Sindbis virus can exploit a host antiviral protein to evade imune surveillance. J. Virol. 2016, 90, 10247–10258. [Google Scholar] [CrossRef]
- Liu, C.H.; Zhou, L.; Chen, G.; Krug, R.M. Battle between influenza A virus and a newly identified antiviral activity of the PARP-containing ZAPL protein. Proc. Natl. Acad. Sci. USA 2015, 112, 14048–14053. [Google Scholar] [CrossRef]
- Sharp, C.P.; Thompson, B.H.; Nash, T.J.; Diebold, O.; Pinto, R.M.; Thorley, L.; Lin, Y.-T.; Sives, S.; Wise, H.; Hendry, S.C.; et al. CpG dinucleotide enrichment in the influenza A virus genome as a live attenuated vaccine development strategy. PLoS Pathog. 2023, 19, e1011357. [Google Scholar] [CrossRef]
- Antzin-Anduetza, I.; Mahiet, C.; Granger, L.A.; Odendall, C.; Swanson, C.M. Increasing the CpG dinucleotide abundance in the HIV-1 genomic RNA inhibits viral replication. Retrovirology 2017, 14, 49. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, J.; Liang, Y.; Nair, V.; Yao, Y.; Cheng, Z. CCCH-type zinc finger antiviral protein mediates antiviral immune response by activating T cells. J. Leukoc. Biol. 2020, 107, 299–307. [Google Scholar] [CrossRef]
- Miyazato, P.; Matsuo, M.; Tan, B.J.Y.; Tokunaga, M.; Katsuya, H.; Islam, S.; Ito, J.; Murakawa, Y.; Satou, Y. HTLV-1 contains a high CG dinucleotide content and is susceptible to the host antiviral protein ZAP. Retrovirology 2019, 16, 38. [Google Scholar] [CrossRef]
- Trus, I.; Udenze, D.; Berube, N.; Wheler, C.; Martel, M.-J.; Gerdts, V.; Karniychuk, U. CpG-Recoding in Zika Virus Genome Causes Host-Age-Dependent Attenuation of Infection With Protection Against Lethal Heterologous Challenge in Mice. Front. Immunol. 2020, 10, 3077. [Google Scholar] [CrossRef]
- Tulloch, F.; Atkinson, N.J.; Evans, D.J.; Ryan, M.D.; Simmonds, P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife 2014, 3, e04531. [Google Scholar] [CrossRef]
- Xie, L.; Lu, B.; Zheng, Z.; Miao, Y.; Liu, Y.; Zhang, Y.; Zheng, C.; Ke, X.; Hu, Q.; Wang, H. The 3C protease of enterovirus A71 counteracts the activity of host zinc-finger antiviral protein (ZAP). J. Gen. Virol. 2018, 99, 73–85. [Google Scholar] [CrossRef]
- Burns, C.C.; Campagnoli, R.; Shaw, J.; Vincent, A.; Jorba, J.; Kew, O. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J. Virol. 2009, 83, 9957–9969. [Google Scholar] [CrossRef]
- Peng, C.; Wyatt, L.S.; Glushakow-Smith, S.G.; Lal-Nag, M.; Weisberg, A.S.; Moss, B. Zinc-finger antiviral protein (ZAP) is a restriction factor for replication of modified vaccinia virus Ankara (MVA) in human cells. PLoS Pathog. 2020, 16, e1008845. [Google Scholar] [CrossRef]
- Chen, E.Q.; Dai, J.; Bai, L.; Tang, H. The efficacy of zinc finger antiviral protein against hepatitis B virus transcription and replication in tansgenic mouse model. Virol. J. 2015, 12, 25. [Google Scholar] [CrossRef]
- Yu, W.; Ji, H.; Long, F.; Chen, S.; He, Q.; Xia, Y.; Cong, C.; Yang, C.; Wei, D.; Huang, F. Inhibition of hepatitis E virus replication by zinc-finger antiviral Protein synergizes with IFN-β. J. Viral Hepat. 2021, 28, 1219–1229. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.S.; Choi, Y.; Son, A.; Park, Y.; Lee, K.M.; Kim, J.; Kim, J.S.; Kim, V.N. The SARS-CoV-2 RNA interactome. Mol. Cell 2021, 81, 2838–2850. [Google Scholar] [CrossRef]
- Tang, A.; Tang, J.; Miao, Q.; Zhu, J.; Guo, H.; Liu, C.; Meng, C.; Li, C.; Chen, Z.; Liu, G. Zinc finger antiviral protein (ZAP) inhibits small ruminant morbillivirus replication in vitro. Vet. Microbiol. 2021, 260, 109163. [Google Scholar] [CrossRef]
- Kypr, J.; Mrazek, J.; Reich, J. Nucleotide composition bias and CpG dinucleotide content in the genomes of HIV and HTLV 1/2. Biochim. Biophys. Acta 1989, 1009, 280–282. [Google Scholar] [CrossRef]
- Ficarelli, M.; Antzin-Anduetza, I.; HughWhite, R.; Firth, A.E.; Sertkaya, H.; Wilson, H.; Neil, S.J.D.; Schulz, R.; Swanson, C.M. CpG dinucleotides inhibit HIV-1 replication through zinc finger antiviral protein (ZAP)-dependent and—Independent mechanisms. J. Virol. 2020, 94, e01337-19. [Google Scholar] [CrossRef]
- Sertkaya, H.; Hidalgo, L.; Ficarelli, M.; Kmiec, D.; Signell, A.W.; Ali, S.; Parker, H.; Wilson, H.; Neil, S.J.; Malim, M.H.; et al. Minimal impact of ZAP on lentiviral vector production and transduction efficiency. Mol. Ther. Methods Clin. Dev. 2021, 23, 147–157. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, J.; Zhou, D.; Yang, K.; Li, B.; Cheng, Z. The CCCH-Type Zinc Finger Antiviral Protein Relieves Immunosuppression of T Cells Induced by Avian Leukosis Virus Subgroup J via the NLP-PKC-δ-NFAT Pathway. J. Virol. 2022, 96, e0134421. [Google Scholar] [CrossRef]
- Pereira-Gómez, M.; Carrau, L.; Fajardo, Á.; Moreno, P.; Moratorio, G. Altering Compositional Properties of Viral Genomes to Design Live-Attenuated Vaccines. Front. Microbiol. 2021, 12, 676582. [Google Scholar] [CrossRef]
- Kozaki, T.; Takahama, M.; Misawa, T.; Matsuura, Y.; Akira, S.; Saitoh, T. Role of zinc-finger anti-viral protein in host defense against Sindbis virus. Int. Immunol. 2015, 27, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Law, L.M.J.; Razooky, B.S.; Li, M.M.H.; You, S.; Jurado, A.; Rice, C.M.; MacDonald, M.R. ZAP’s stress granule localization is correlated with its antiviral activity and induced by virus replication. PLOS Pathog. 2019, 15, e1007798. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Q.; Wang, X.; Gao, G. The short form of the zinc finger antiviral protein inhibits influenza A virus protein expression and is antagonized by the virus-encoded NS1. J. Virol. 2017, 91, e01909-16. [Google Scholar]
- Xuan, Y.; Gong, D.; Qi, J.; Han, C.; Deng, H.; Gao, G. ZAP inhibits murine gammaherpesvirus 68 ORF64 expression and is antagonized by RTA. J. Virol. 2013, 87, 2735–2743. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, E.; Wise, H.M.; Zhang, H.; Lee, L.N.; Atkinson, N.J.; Nicol, M.Q.; Highton, A.J.; Klenerman, P.; Beard, P.M.; Dutia, B.M.; et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. eLife 2016, 5, e12735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, W.; Chen, S.; Yuan, Z.; Yi, Z. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antivir. Res. 2021, 185, 104974. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Sun, Z.; Yu, L.; Shi, D.; Zhu, M.; Yao, H.; Li, L. Interactome Analysis of the Nucleocapsid Protein of SARS-CoV-2 Virus. Pathogens 2021, 10, 1155. [Google Scholar] [CrossRef] [PubMed]
- Kamel, W.; Noerenberg, M.; Cerikan, B.; Chen, H.; Järvelin, A.I.; Kammoun, M.; Lee, J.Y.; Shuai, N.; Garcia-Moreno, M.; Andrejeva, A.; et al. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol. Cell 2021, 81, 2851–2867. [Google Scholar] [CrossRef]
- Afrasiabi, A.; Alinejad-Rokny, H.; Khosh, A.; Rahnama, M.; Lovell, N.; Xu, Z.; Ebrahimi, D. The low abundance of CpG in the SARS-CoV-2 genome is not an evolutionarily signature of ZAP. Sci. Rep. 2022, 12, 2420. [Google Scholar] [CrossRef]
- Forni, D.; Pozzoli, U.; Cagliani, R.; Clerici, M.; Sironi, M. Dinucleotide biases in RNA viruses that infect vertebrates or invertebrates. Microbiol. Spectr. 2023, 11, e0252923. [Google Scholar] [CrossRef]


| Virus Family | Effect of ZAP on Viruses | Cofactors Required by ZAP | Associated Immune Pathways | References | |
|---|---|---|---|---|---|
| Togaviridae | Sindbis virus (SINV) |
| TRIM25 | Type I IFN | [7,15,16,18,20,38,108,109] |
| Semliki Forest virus (SFV) |
| Not measured | Not measured | [7,15] | |
| Ross River virus (RRV) |
| Not measured | Not measured | [7] | |
| Orthomyxoviridae | Influenza A virus |
| Not measured |
| [40,103,110,111] |
| Retroviridae | Moloney and murine leukemia virus (MMLV or MuLV) |
|
|
| [13,15,30,103] |
| Human immunodeficiency virus type 1 (HIV-1) |
|
| Not measured | [22,25,52,55,112] | |
| Avian leukosis virus subgroup J (ALV-J) |
| Not measured | Not measured | [113] | |
| Human T-lymphotropic virus type 1 (HTLV-1) |
| Not measured | Not measured | [114] | |
| Flaviviridae | Japanese encephalitis virus (JEV) |
|
|
| [10] |
| Zika virus (ZIKV) |
| Not measured | Not measured | [26,115] | |
| Filoviridae | Ebola virus (EBOV) |
| TRIM25 | Type I IFN | [6,12,16] |
| Marburg virus (MARV) |
| Not measured | Not measured | [6] | |
| Picornaviridae | Coxsackievirus B3 |
| Not measured | Not measured | [14] |
| Echovirus 7 (E7) |
|
| Not measured | [60,89,116] | |
| Enterovirus A71 (EV-A71) |
| Not measured | Not measured | [117] | |
| Poliovirus |
| Not measured | Not measured | [118] | |
| Poxviridae | Vaccinia virus Ankara (MVA) |
| Not measured | Not measured | [119] |
| Hepadnaviridae | Hepatitis B virus |
| Not measured | Not measured | [16,31,120] |
| Hepeviridae | Hepatitis E virus |
| Not measured | IRF3 signaling | [121] |
| Herpesviridae | Murine gammaherpesvirus 68 |
| Not measured | Not measured | [33] |
| Human cytomegalovirus (HCMV) |
| TRIM25 | RIG-I and IRF3 signaling | [70] | |
| Coronaviridae | Severe acute respiratory syndrome Enterovirus A71 s 2 (SARS-CoV-2) |
| TRIM25KHNYN | Type I and II IFN | [24,25,51,122] |
| Paramyxoviridae | Sendai virus |
| Not measured | Type I IFN; IRF3 | [11,16] |
| Small ruminant morbillivirus (SRMV) |
| Not measured | Not measured | [123] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Andrade, K.Q.; Cirne-Santos, C.C. Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families. Pathogens 2023, 12, 1461. https://doi.org/10.3390/pathogens12121461
de Andrade KQ, Cirne-Santos CC. Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families. Pathogens. 2023; 12(12):1461. https://doi.org/10.3390/pathogens12121461
Chicago/Turabian Stylede Andrade, Kívia Queiroz, and Claudio Cesar Cirne-Santos. 2023. "Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families" Pathogens 12, no. 12: 1461. https://doi.org/10.3390/pathogens12121461
APA Stylede Andrade, K. Q., & Cirne-Santos, C. C. (2023). Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families. Pathogens, 12(12), 1461. https://doi.org/10.3390/pathogens12121461







