Effect of a Depolymerase Encoded by Phage168 on a Carbapenem-Resistant Klebsiella pneumoniae and Its Biofilm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Purification of Phage
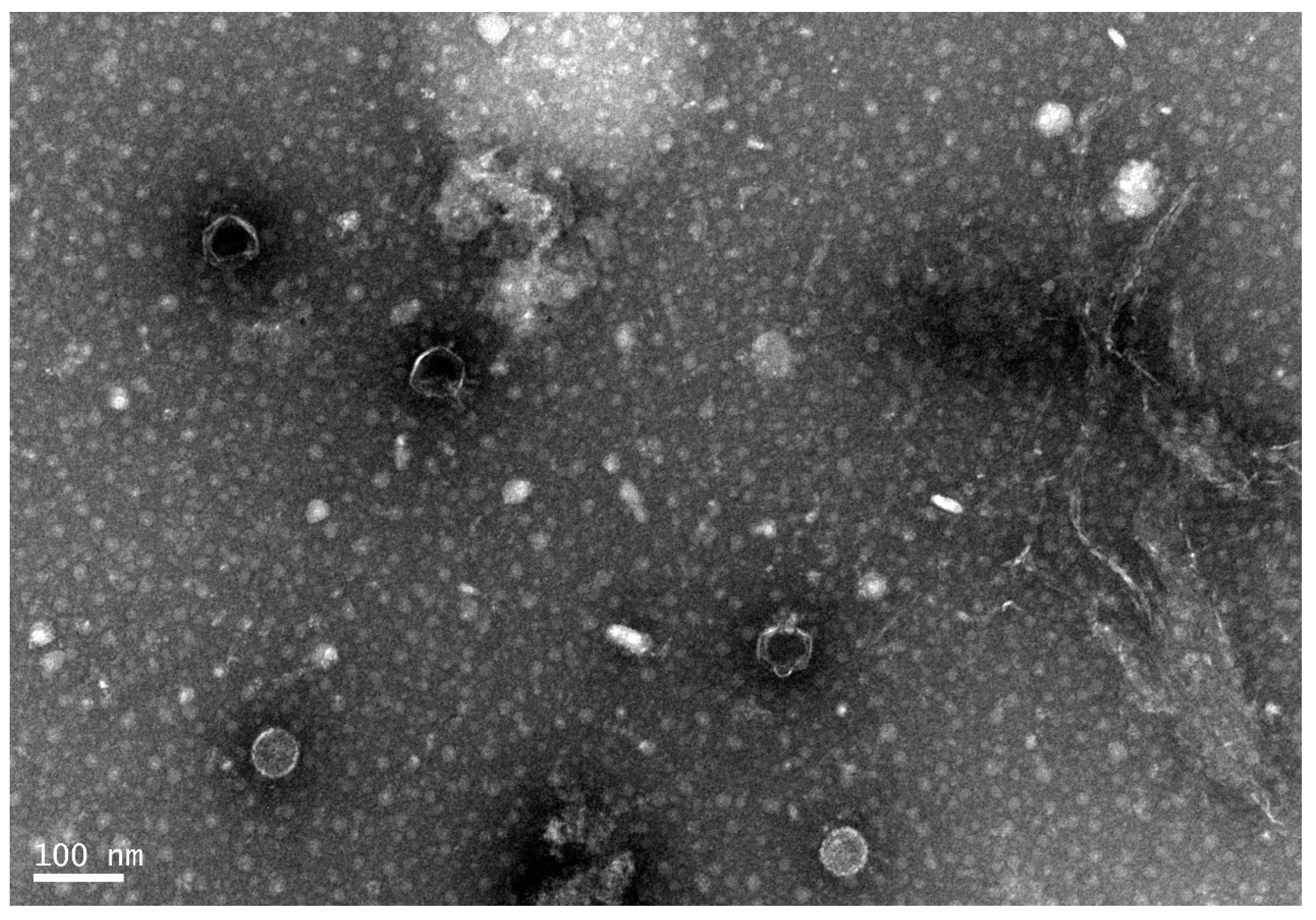
2.2. Transmission Electron Microscopy
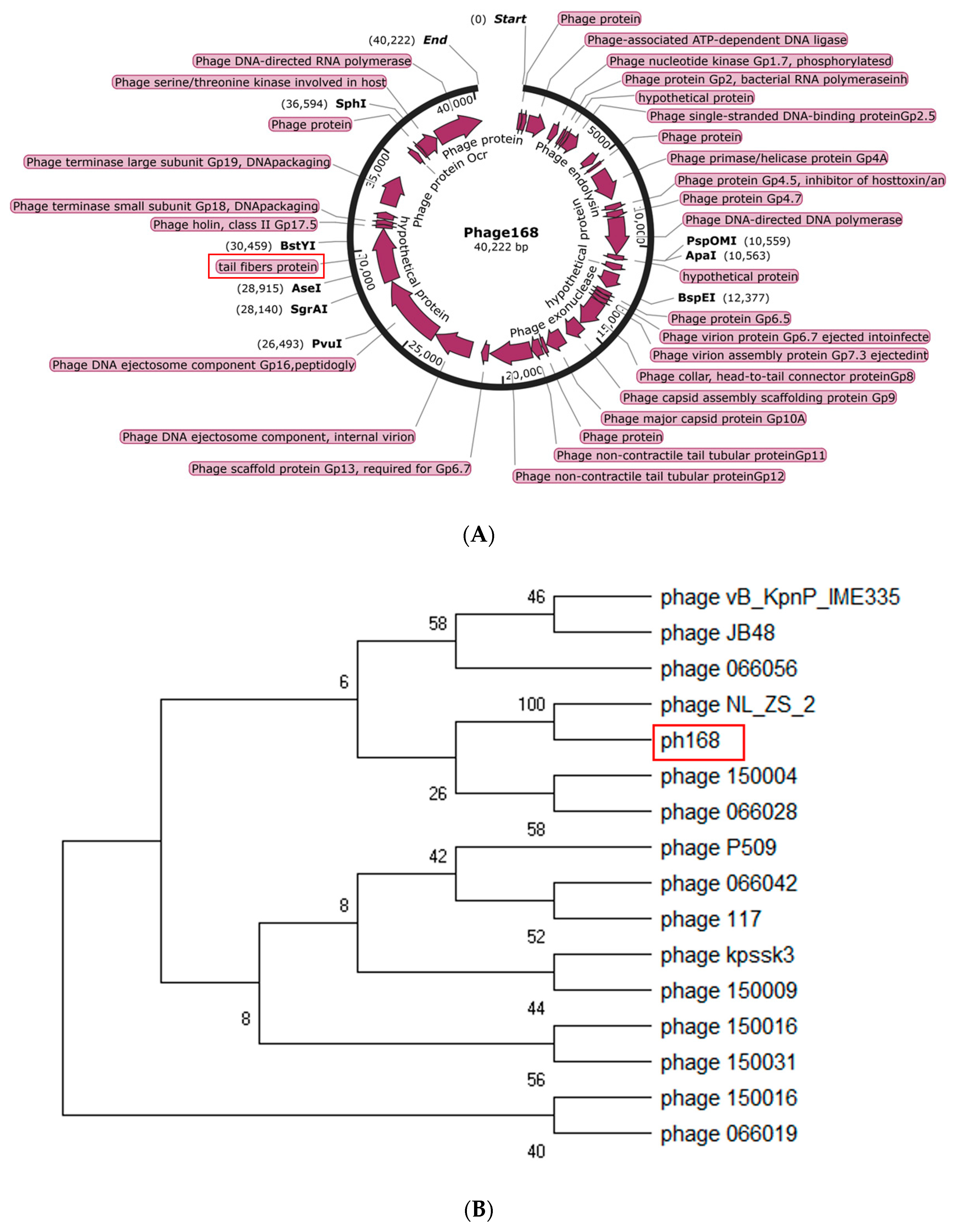
2.3. Genomic DNA Sequencing and Annotation
2.4. Phylogenetic Analysis
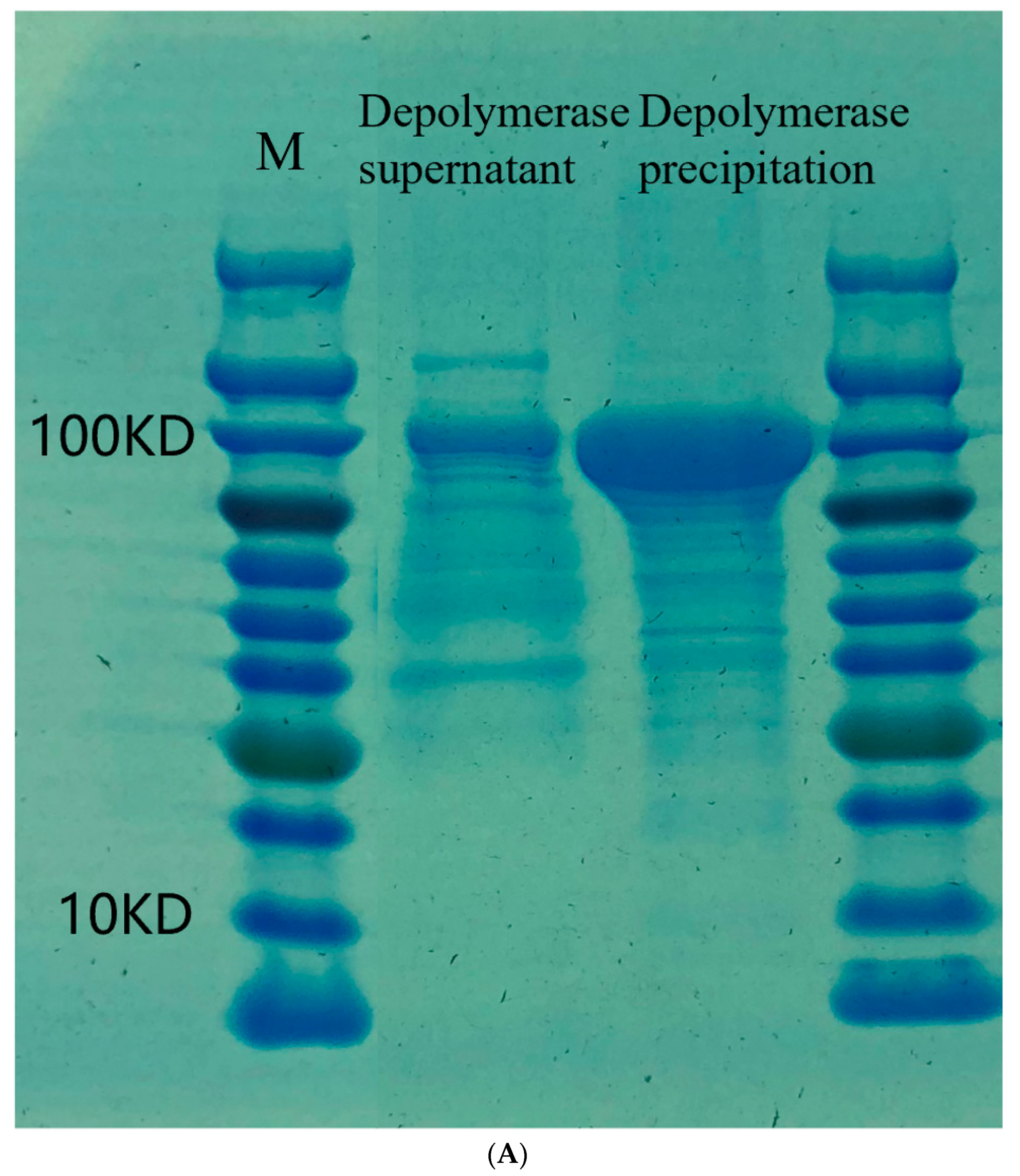
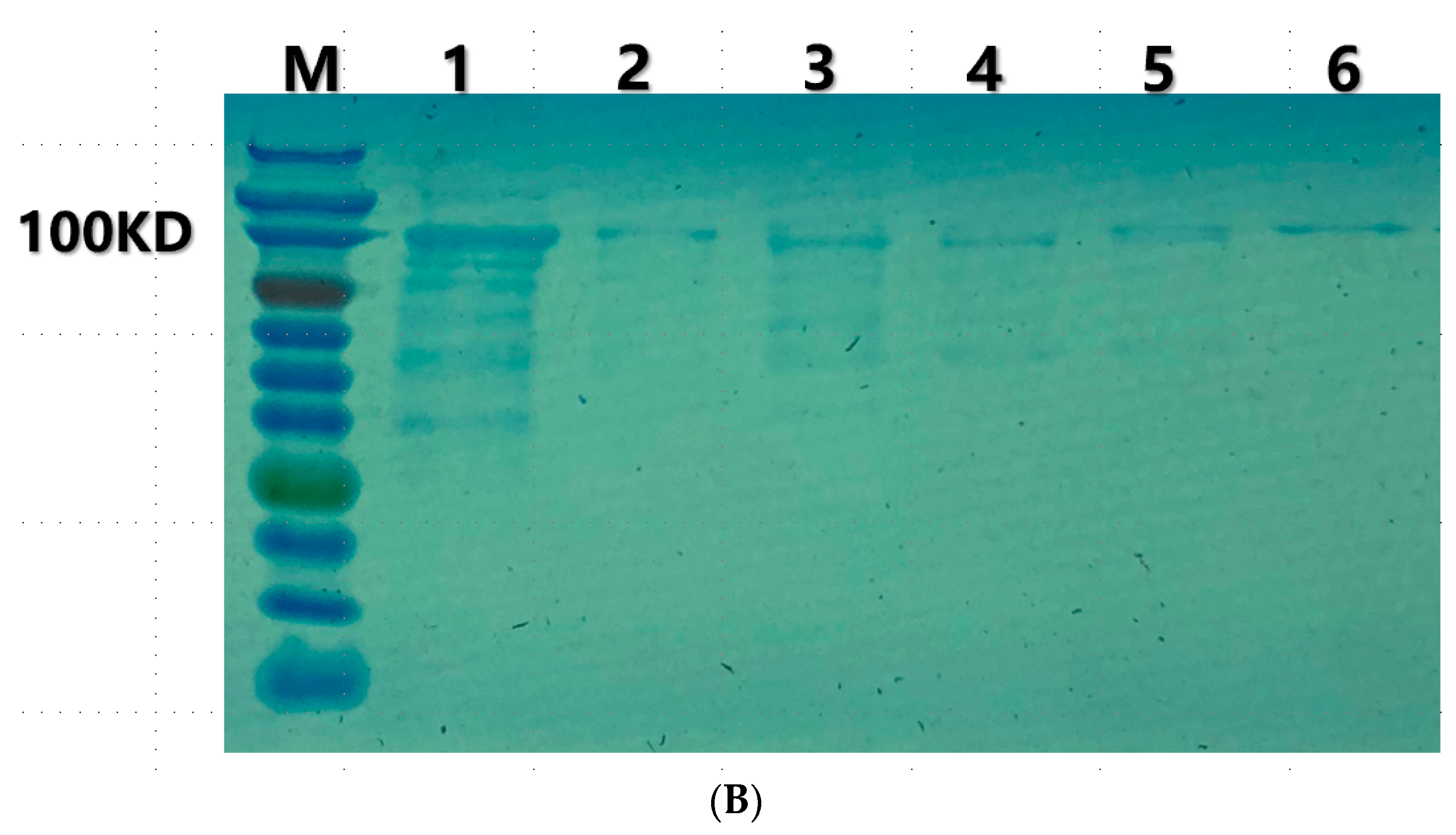
2.5. Cloning, Expression and Purification of the Recombinant Depolymerase
2.6. Depolymerization Activity Assay
2.7. Screening of Biofilm Producing Strains in CRKP
2.7.1. Construction of Biofilm
2.7.2. Semi-Quantitative Detection of Biofilms
2.7.3. Effects of Depolymerase on Formed CRKP Biofilms
2.7.4. Antibiofilm Activity Combined with Antibiotics
2.7.5. Serum Killing Assay
2.8. Statistical Analysis
3. Results
3.1. General Microbiological Characteristics of Phage168
3.2. Analysis of the Genome of Phage168
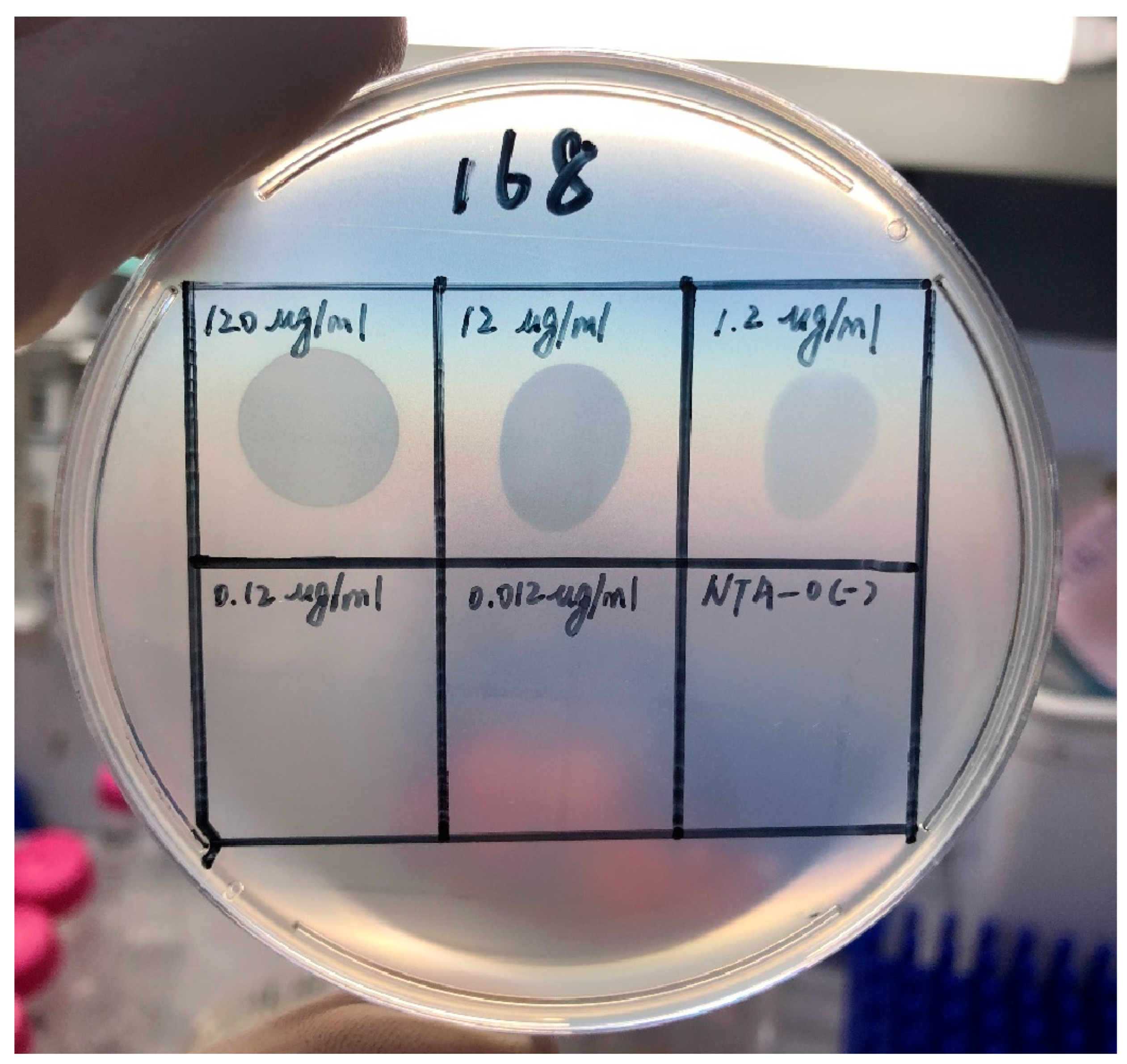
3.3. Depolymerization Activity Assay
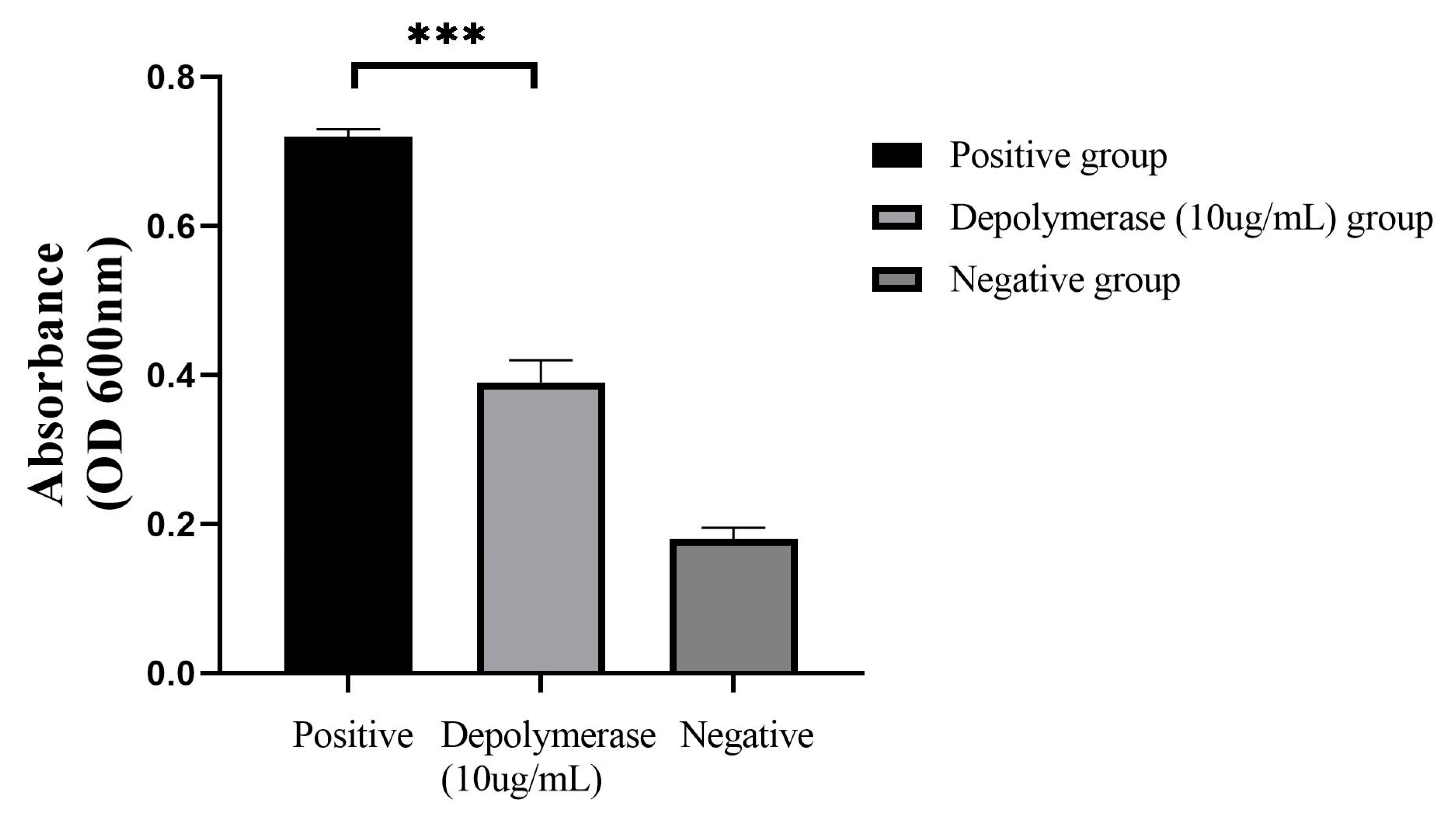
3.4. Depolymerase Could Destroy the CRKP-Formed Biofilm
3.5. The Effect of Depolymerase Can Be Combined with That of Polymyxin B on CRKP Biofilm
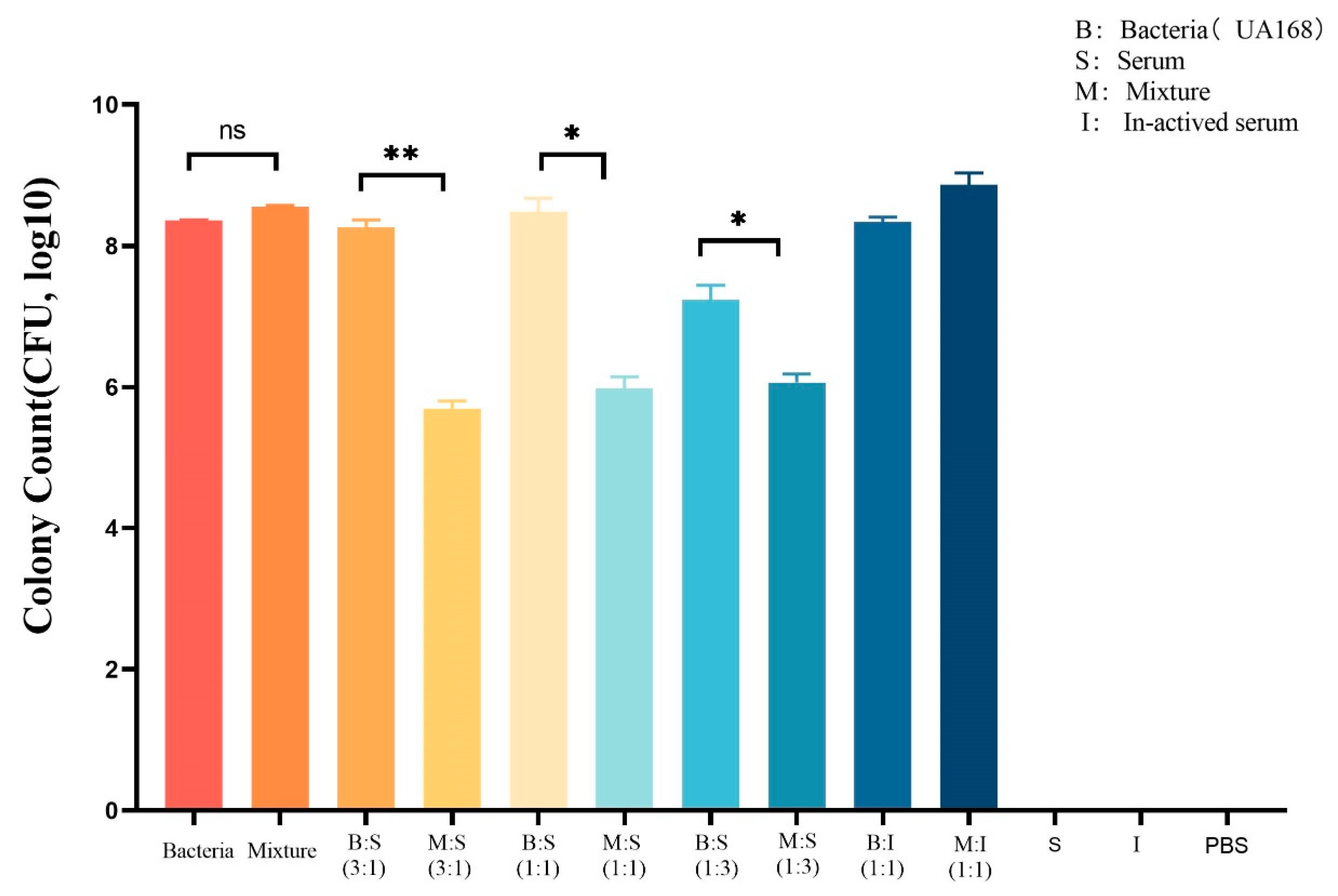
3.6. Depolymerase Can Enhance the Serum-Mediated Bactericidal Effect on CRKP
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, F.; Guo, Y.; Zhu, D.; Wang, F.; Jiang, X.; Xu, Y.C.; Zhang, X.; Zhang, Z.; Ji, P.; Xie, Y.; et al. CHINET surveillance of bacterial resistance across tertiary hospitals in 2019. Chin. J. Infect. Chemother. 2020, 20, 233–243. [Google Scholar]
- Qiao, F.; Huang, W.; Zong, Z. Significance and development trend of multi-drug resistant bacteria infection prevention and control. Chin. Nurs. Manag. 2019, 19, 6. [Google Scholar]
- Hsieh, P.F.; Lin, T.L.; Lee, C.Z.; Tsai, S.F.; Wang, J.T. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2008, 197, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Amrani, A.; Hamed, M.B.B.; Talebbendiab, F.M. Association study between some renin-angiotensin system gene variants and essential hypertension in a sample of Algerian population: Case control study. Ann. Biol. Clin. 2015, 73, 557–563. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Cortés, G.; Borrell, N.; de Astorza, B.; Gómez, C.; Sauleda, J.; Albertí, S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 2002, 70, 2583–2590. [Google Scholar] [CrossRef] [PubMed]
- Ciurana, B.; Tomás, J.M. Role of lipopolysaccharide and complement in susceptibility of Klebsiella pneumoniae to nonimmune serum. Infect. Immun. 1987, 55, 2741–2746. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3, 223–247. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef]
- Li, X.-H.; Lee, J.-H. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P. A Novel Tail-Associated O91-Specific Polysaccharide Depolymerase from a Podophage Reveals Lytic Efficacy of Shiga Toxin-Producing Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e00145-20. [Google Scholar] [CrossRef]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Choi, Y.-J.; Koh, W.S.; Moon, K.S.; Kang, S.H. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob. Agents Chemother. 2014, 58, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Topka, G.; Bloch, S.; Nejman-Faleńczyk, B.; Gąsior, T.; Jurczak-Kurek, A.; Necel, A.; Dydecka, A.; Richert, M.; Węgrzyn, G.; Węgrzyn, A. Characterization of bacteriophage vB-EcoS-95, isolated from urban sewage and revealing extremely rapid lytic development. Front. Microbiol. 2019, 9, 3326. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Luo, T.; Yang, Y.; Dong, D.; Wang, R.; Wang, Y.; Xu, M.; Guo, X.; Hu, F.; He, P. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front. Microbiol. 2018, 8, 2659. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.; Parasion, S.; Rutyna, P.; Mizak, L.; Gryko, R.; Niemcewicz, M.; Olender, A.; Łobocka, M. Isolation of bacteriophages and their application to control Pseudomonas aeruginosa in planktonic and biofilm models. Resn Microbiol. 2016, 168, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, M.; Liu, Q.; Liu, Y.; Wang, Q.; Duan, X.; Liu, L.; Jiang, Y.; Shao, H.; Guo, C. Genome analysis of two novel lytic vibrio maritimus phages isolated from the coastal surface seawater of Qingdao, China. Curr. Microbiol. 2019, 76, 1225–1233. [Google Scholar] [CrossRef]
- Walker, J.M. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol. Biol. 1994, 32, 5–8. [Google Scholar] [PubMed]
- Kovalchuk, V.P.; Nazarchuk, O.A.; Burkot, V.M.; Fomina, N.S.; Prokopchuk, Z.M.; Dobrovanov, O. Biofilm forming activity of non-fermenting gram-negative bacteria. Wiad. Lek. 2021, 74, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Y.; Bao, R.; Zhu, T.; Tan, D.; Hu, B. Isolation and Characterization of Novel Phages Targeting Pathogenic Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 2021, 11, 792305. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, R.A.; Bamert, R.S.; Belousoff, M.J.; Short, F.L.; Barlow, C.K.; Pickard, D.J.; Wilksch, J.J.; Schittenhelm, R.B.; Strugnell, R.A.; Dougan, G.; et al. Mechanistic Insights into the Capsule-Targeting Depolymerase from a Klebsiella pneumoniae Bacteriophage. Microbiol. Spectr. 2021, 9, e0102321. [Google Scholar] [CrossRef] [PubMed]
- Sahly, H.; Podschun, R.; Ullmann, U. Klebsiella infections in the immunocompromised host. Adv. Exp. Med. Biol. 2000, 479, 237–249. [Google Scholar] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: The WHO priority list of antibioticresistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cao, F.; Wang, X.; Wang, L.; Li, Z.; Che, J.; Wang, L.; Li, X.; Cao, Z.; Zhang, J.; Jin, L.; et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed. Res. Int. 2015, 2015, 752930. [Google Scholar] [CrossRef]
- Singla, S.; Harjai, K.; Katare, O.P.; Chhibber, S. Bacteriophage-loaded nanostructured lipid carrier: Improved pharmacokinetics mediates effective resolution of Klebsiella pneumoniae-induced lobar pneumonia. J. Infect. Dis. 2015, 212, 325–334. [Google Scholar] [CrossRef]
- Aleshkin, A.V.; Ershova, O.N.; Volozhantsev, N.V.; Svetoch, E.A.; Popova, A.V.; Rubalskii, E.O.; Borzilov, A.I.; Aleshkin, V.A.; Afanas’ Ev, S.S.; Karaulov, A.V.; et al. Phagebiotics in treatment and prophylaxis of healthcare-associated infections. Bacteriophage 2016, 6, e1251379. [Google Scholar] [CrossRef]
- Chadha, P.; Katare, O.P.; Chhibber, S. Liposome loaded phage cocktail: Enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 2017, 43, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and Tolerance of the Phage Endolysin-Based Candidate Drug SAL200 after a Single Intravenous Administration among Healthy Volunteers. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Petrovic Fabijan, A.; Lin, R.C.Y.; Ho, J.; Maddocks, S.; Ben Zakour, N.L.; Iredell, J.R.; Westmead Bacteriophage Therapy Team. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 2020, 5, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P.-J.; Krylov, V.N.; Noben, J.-P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Steven, A.; Trus, B.; Maizel, J.; Unser, M.; Parry, D.; Wall, J.; Hainfeld, J.; Studier, F. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 1988, 200, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef]
- Murofushi, Y.; Villena, J.; Morie, K.; Kanmani, P.; Tohno, M.; Shimazu, T.; Aso, H.; Suda, Y.; Hashiguchi, K.; Saito, T.; et al. The toll-like receptor family protein RP105/MD1 complex is involved in the immunoregulatory effect of exopolysaccharides from Lactobacillus plantarum N14. Mol. Immunol. 2015, 64, 63–75. [Google Scholar] [CrossRef]
- Kundukad, B.; Schussman, M.; Yang, K.; Seviour, T.; Yang, L.; Rice, S.A.; Kjelleberg, S.; Doyle, P.S. Mechanistic action of weak acid drugs on biofilms. Sci. Rep. 2017, 7, 4783. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Boudarel, H.; Mathias, J.-D.; Blaysat, B.; Grédiac, M. Towards standardized mechanical characterization of microbial biofilms: Analysis and critical review. NPJ Biofilms Microbiomes 2018, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, P.; Niu, W.; Yuan, X.; Liu, H.; Huang, Y.; An, X.; Fan, H.; Zhangxiang, L.; Mi, L.; et al. Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Res. Microbiol. 2019, 170, 156–164. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Briers, Y.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Role of the Pre-neck Appendage Protein (Dpo7) from Phage vB_SepiSphiIPLA7 as an Anti-biofilm Agent in Staphylococcal Species. Front. Microbiol. 2015, 6, 1315. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Harjai, K.; Chhibber, S. Aeromonas punctata derived depolymerase improves susceptibility of Klebsiella pneumoniae biofilm to gentamicin. BMC Microbiol. 2015, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Harjai, K.; Chhibber, S. Depolymerase improves gentamicin efficacy during Klebsiella pneumoniae induced murine infection. BMC Infect. Dis. 2014, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Domenico, P.; Salo, R.J.; Cross, A.S.; Cunha, B.A. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 1994, 62, 4495–4499. [Google Scholar] [CrossRef]
- Lin, T.-L.; Hsieh, P.-F.; Huang, Y.-T.; Lee, W.-C.; Tsai, Y.-T.; Su, P.-A.; Pan, Y.-J.; Hsu, C.-R.; Wu, M.-C.; Wang, J.-T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: Implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef]
- Oliveira, H.; Mendes, A.; Fraga, A.G.; Ferreira, A.; Pimenta, A.I.; Mil-Homens, D.; Fialho, A.M.; Pedrosa, J.; Azeredo, J. K2 Capsule Depolymerase Is Highly Stable, Is Refractory to Resistance, and Protects Larvae and Mice from Acinetobacter baumannii Sepsis. Appl. Env. Microbiol. 2019, 85, e00934-19. [Google Scholar] [CrossRef]
- Liu, Y.; Mi, Z.; Mi, L.; Huang, Y.; Li, P.; Liu, H.; Yuan, X.; Niu, W.; Jiang, N.; Bai, C.; et al. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 2019, 7, e6173. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Łątka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses 2016, 8, 324. [Google Scholar] [CrossRef]
- Oliveira, H.; Pinto, G.; Mendes, B.; Dias, O.; Hendrix, H.; Akturk, E.; Noben, J.-P.; Gawor, J.; Łobocka, M.; Lavigne, R.; et al. A Tailspike with Exopolysaccharide Depolymerase Activity from a New Providencia stuartii Phage Makes Multidrug-Resistant Bacteria Susceptible to Serum-Mediated Killing. Appl. Env. Microbiol. 2020, 86, e00073-20. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.F.; Lin, H.H.; Lin, T.L.; Chen, Y.Y.; Wang, J.T. Two T7-like Bacteriophages, K5-2 and K5-4, Each Encodes Two Capsule Depolymerases: Isolation and Functional Characterization. Sci. Rep. 2017, 7, 4624. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Paff, M.L.; Molineux, I.J.; Bull, J.J. Therapeutic Application of Phage Capsule Depolymerases against K1, K5, and K30 Capsulated E. coli in Mice. Front. Microbiol. 2017, 8, 2257. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, K.; Gutiérrez, D.; Latka, A.; Boeckaerts, D.; Drulis-Kawa, Z.; Criel, B.; Gerstmans, H.; Safaan, A.; Khairalla, A.S.; Gaber, Y.; et al. The Specific Capsule Depolymerase of Phage PMK34 Sensitizes Acinetobacter baumannii to Serum Killing. Antibiotics 2022, 11, 677. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 84–99. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Pu, B.; Qin, J.; Xiang, J. Effect of a Depolymerase Encoded by Phage168 on a Carbapenem-Resistant Klebsiella pneumoniae and Its Biofilm. Pathogens 2023, 12, 1396. https://doi.org/10.3390/pathogens12121396
Sun X, Pu B, Qin J, Xiang J. Effect of a Depolymerase Encoded by Phage168 on a Carbapenem-Resistant Klebsiella pneumoniae and Its Biofilm. Pathogens. 2023; 12(12):1396. https://doi.org/10.3390/pathogens12121396
Chicago/Turabian StyleSun, Xu, Bingchun Pu, Jinhong Qin, and Jun Xiang. 2023. "Effect of a Depolymerase Encoded by Phage168 on a Carbapenem-Resistant Klebsiella pneumoniae and Its Biofilm" Pathogens 12, no. 12: 1396. https://doi.org/10.3390/pathogens12121396
APA StyleSun, X., Pu, B., Qin, J., & Xiang, J. (2023). Effect of a Depolymerase Encoded by Phage168 on a Carbapenem-Resistant Klebsiella pneumoniae and Its Biofilm. Pathogens, 12(12), 1396. https://doi.org/10.3390/pathogens12121396





