Single Amino Acid Polymorphisms in the Fasciola hepatica Carboxylesterase Type B Gene and Their Potential Role in Anthelmintic Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Parasite Strains
2.2. Parasite Material
2.3. DNA Extraction
2.4. PCR Conditions
2.5. Sanger Amplicon Sequencing
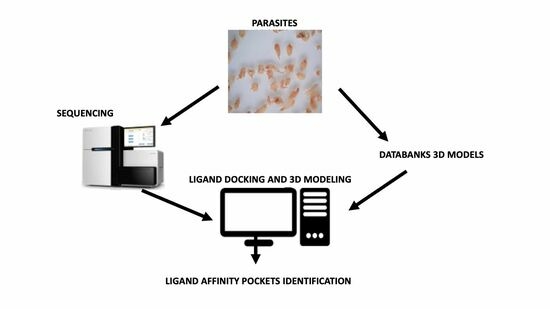
2.6. Protein Sequences and 3D Models
2.7. Protein—Ligand Docking 3D Modeling
3. Results
3.1. Parasites Samples
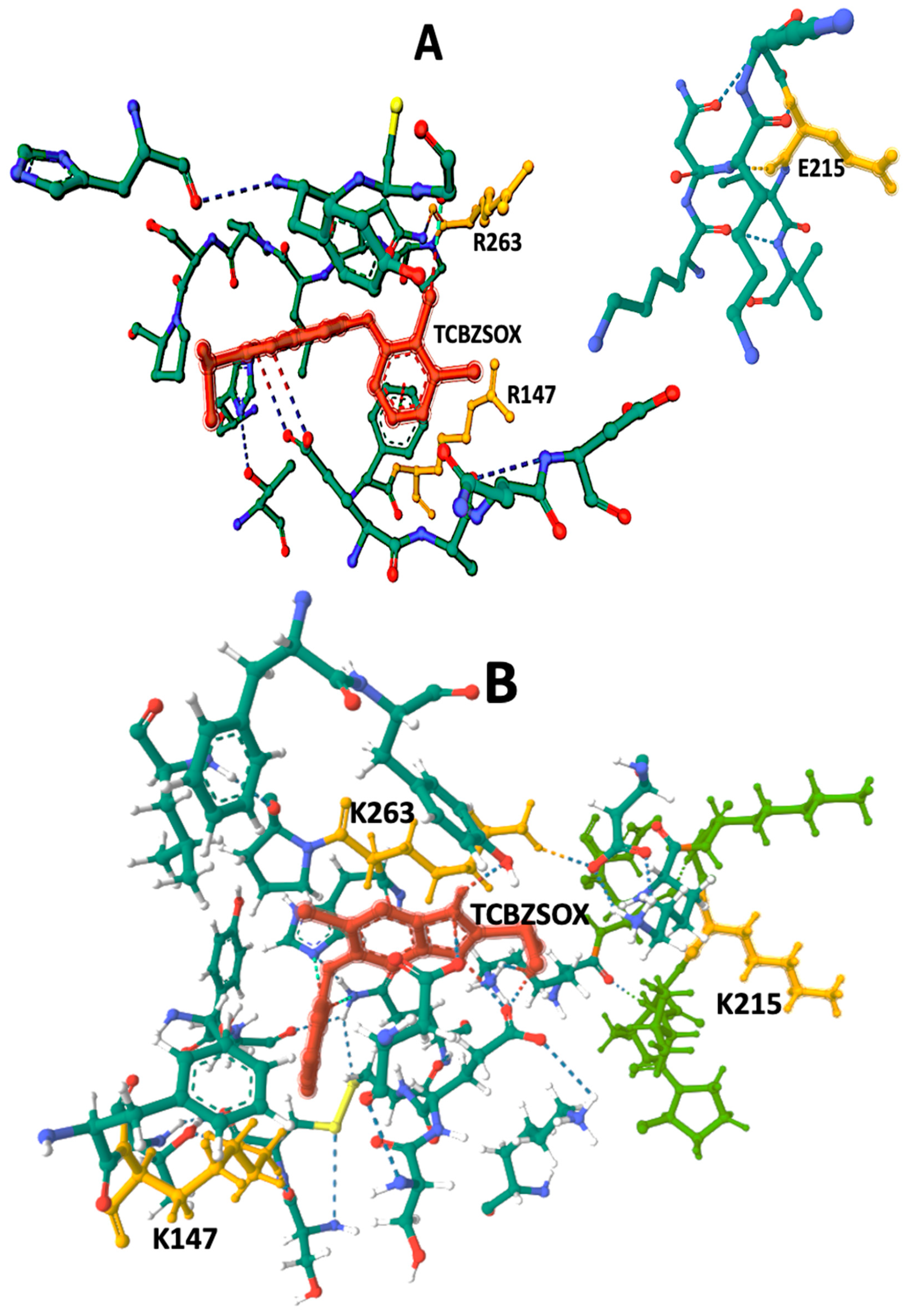
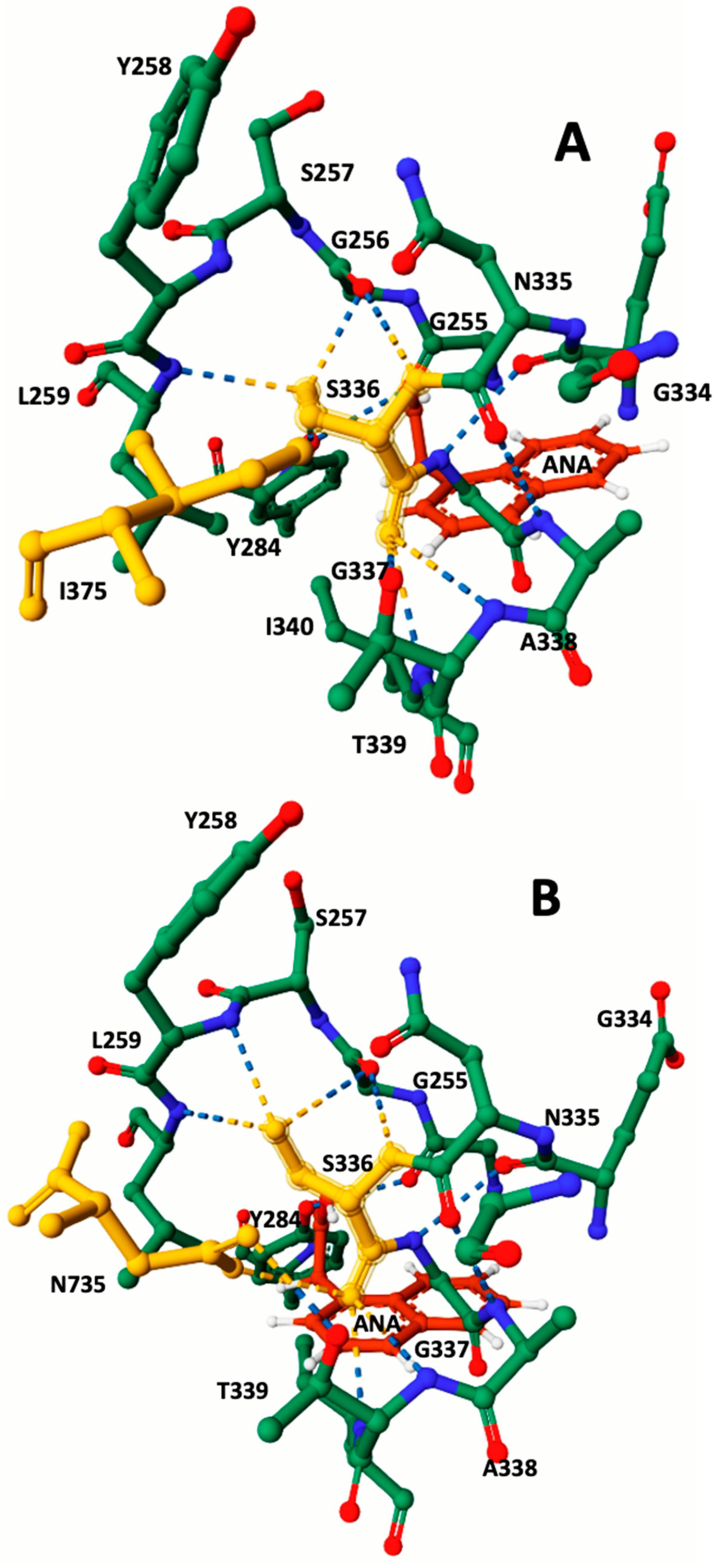
3.2. Protein 3D Modeling and Protein–Ligand Docking Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fissiha, W.; Kinde, M.Z. Anthelmintic Resistance and Its Mechanism: A Review. Infect. Drug Resist. 2021, 14, 5403–5410. [Google Scholar] [CrossRef] [PubMed]
- Brennan, G.P.; Fairweather, I.; Trudgett, A.; Hoey, E.; McCoy; McConville, M.; Meaney, M.; Robinson, M.; McFerran, N.; Ryan, L.; et al. Understanding Triclabendazole Resistance. Exp. Mol. Pathol. 2007, 82, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.M.; Cabada, M.M. Recent Developments in the Epidemiology, Diagnosis, and Treatment of Fasciola Infection. Curr. Opin. Infect. Dis. 2018, 31, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Vivas, R.I.; Grisi, L.; Pérez De León, A.A.; Silva Villela, H.; Torres-Acosta, J.F.D.J.; Fragoso Sánchez, H.; Romero Salas, D.; Rosario Cruz, R.; Saldierna, F.; García Carrasco, D. Potential Economic Impact Assessment for Cattle Parasites in Mexico. Review. Rev. Mex. Cienc. Pecu. 2017, 8, 61. [Google Scholar] [CrossRef]
- Zumaquero-Ríos, J.L.; Sarracent-Pérez, J.; Rojas-García, R.; Rojas-Rivero, L.; Martínez-Tovilla, Y.; Valero, M.A.; Mas-Coma, S. Fascioliasis and Intestinal Parasitoses Affecting Schoolchildren in Atlixco, Puebla State, Mexico: Epidemiology and Treatment with Nitazoxanide. PLoS Negl. Trop. Dis. 2013, 7, e2553. [Google Scholar] [CrossRef]
- Sánchez Vega, J.T.; Morales Galicia, A.E.; Hernández López, R.; Navez Valle, A.; Morales Reyes, E.G.; Sánchez Aguilar, D.I.; Tapia Castor, A.C.; Coquis Téllez, B.; Hernández Covarrubias, R.I. Panorama General de Las Helmintiasis Extraintestinales En México Durante Las Últimas Dos Décadas. Lux Méd. 2022, 17, 5–13. [Google Scholar] [CrossRef]
- Fairweather, I. Raising the Bar on Reporting ‘Triclabendazole Resistance’. Vet. Rec. 2011, 168, 514–515. [Google Scholar] [CrossRef]
- Matoušková, P.; Vokřál, I.; Lamka, J.; Skálová, L. The Role of Xenobiotic-Metabolizing Enzymes in Anthelmintic Deactivation and Resistance in Helminths. Trends Parasitol. 2016, 32, 481–491. [Google Scholar] [CrossRef]
- Mordvinov, V.; Pakharukova, M. Xenobiotic-Metabolizing Enzymes in Trematodes. Biomedicines 2022, 10, 3039. [Google Scholar] [CrossRef]
- Wheelock, C.E.; Shan, G.; Ottea, J. Overview of Carboxylesterases and Their Role in the Metabolism of Insecticides. J. Pestic. Sci. 2005, 30, 75–83. [Google Scholar] [CrossRef]
- Hatfield, M.J.; Umans, R.A.; Hyatt, J.L.; Edwards, C.C.; Wierdl, M.; Tsurkan, L.; Taylor, M.R.; Potter, P.M. Carboxylesterases: General Detoxifying Enzymes. Chem.-Biol. Interact. 2016, 259, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Bhatt, K.; Huang, Y.; Lin, Z.; Chen, S. Esterase Is a Powerful Tool for the Biodegradation of Pyrethroid Insecticides. Chemosphere 2020, 244, 125507. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, R.D.; Campbell, P.M.; Ollis, D.L.; Cheah, E.; Russell, R.J.; Oakeshott, J.G. A Single Amino Acid Substitution Converts a Carboxylesterase to an Organophosphorus Hydrolase and Confers Insecticide Resistance on a Blowfly. Proc. Natl. Acad. Sci. USA 1997, 94, 7464–7468. [Google Scholar] [CrossRef] [PubMed]
- Cruse, C.; Moural, T.W.; Zhu, F. Dynamic Roles of Insect Carboxyl/Cholinesterases in Chemical Adaptation. Insects 2023, 14, 194. [Google Scholar] [CrossRef]
- Hopkins, D.H.; Fraser, N.J.; Mabbitt, P.D.; Carr, P.D.; Oakeshott, J.G.; Jackson, C.J. Structure of an Insecticide Sequestering Carboxylesterase from the Disease Vector Culex quinquefasciatus: What Makes an Enzyme a Good Insecticide Sponge? Biochemistry 2017, 56, 5512–5525. [Google Scholar] [CrossRef]
- Wei, P.; Demaeght, P.; De Schutter, K.; Grigoraki, L.; Labropoulou, V.; Riga, M.; Vontas, J.; Nauen, R.; Dermauw, W.; Van Leeuwen, T. Overexpression of an Alternative Allele of Carboxyl/Choline Esterase 4 (CCE04) of Tetranychus urticae Is Associated with High Levels of Resistance to the Keto-enol Acaricide Spirodiclofen. Pest Manag. Sci. 2020, 76, 1142–1153. [Google Scholar] [CrossRef]
- Devonshire, A.L.; Field, L.M.; Foster, S.P.; Moores, G.D.; Williamson, M.S.; Blackman, R.L. The Evolution of Insecticide Resistance in the Peach–Potato Aphid, Myzus Persicae. Phil. Trans. R. Soc. Lond. B 1998, 353, 1677–1684. [Google Scholar] [CrossRef]
- Scarcella, S.; Solana, M.V.; Fernandez, V.; Lamenza, P.; Ceballos, L.; Solana, H. Increase of Gluthatione S-Transferase, Carboxyl Esterase and Carbonyl Reductase in Fasciola Hepatica Recovered from Triclabendazole Treated Sheep. Mol. Biochem. Parasitol. 2013, 191, 63–65. [Google Scholar] [CrossRef]
- Pedroza-Gómez, Y.J.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Scarcella, S.; Reynaud, E.; Sanchez-Carbente, M.D.R.; Narváez-Padilla, V.; Miranda-Miranda, E. Transcriptome-Based Identification of a Functional Fasciola Hepatica Carboxylesterase B. Pathogens 2021, 10, 1454. [Google Scholar] [CrossRef]
- Miranda-Miranda, E.; Scarcella, S.; Reynaud, E.; Narváez-Padilla, V.; Neira, G.; Mera-y-Sierra, R.; Aguilar-Díaz, H.; Cossio-Bayugar, R. A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola Hepatica Carboxylesterase B. Genes 2022, 13, 1899. [Google Scholar] [CrossRef]
- Miranda-Miranda, E.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Narváez-Padilla, V.; Sachman-Ruíz, B.; Reynaud, E. Transcriptome Assembly Dataset of Anthelmintic Response in Fasciola Hepatica. Data Brief 2021, 35, 106808. [Google Scholar] [CrossRef] [PubMed]
- Scarcella, S.; Miranda-Miranda, E.; Cossío-Bayúgar, R.; Ceballos, L.; Fernandez, V.; Solana, H. Increase of Carboxylesterase Activity in Fasciola Hepatica Recovered from Triclabendazole Treated Sheep. Mol. Biochem. Parasitol. 2012, 185, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Calvani, N.E.D.; Windsor, P.A.; Bush, R.D.; Šlapeta, J. Scrambled Eggs: A Highly Sensitive Molecular Diagnostic Workflow for Fasciola Species Specific Detection from Faecal Samples. PLoS Negl. Trop. Dis. 2017, 11, e0005931. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Rusell, D.W. (Eds.) Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 978-0-87969-577-4. [Google Scholar]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein–Ligand Binding Site Recognition Using Complementary Binding-Specific Substructure Comparison and Sequence Profile Alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef]
- Ibarra-Velarde, F.; Vera-Montenegro, Y.; Munguia-Xochihua, J. Capítulo 9. Epidemiología de La Fasciolosis Animal y Humana. In Epidemiología de Enfermedades Parasitarias en Animales Domésticos; Compact Disc CD-ROM: Mexico City, Mexico, 2011; pp. 137–155. ISBN 978-607-00-4015-3. [Google Scholar]
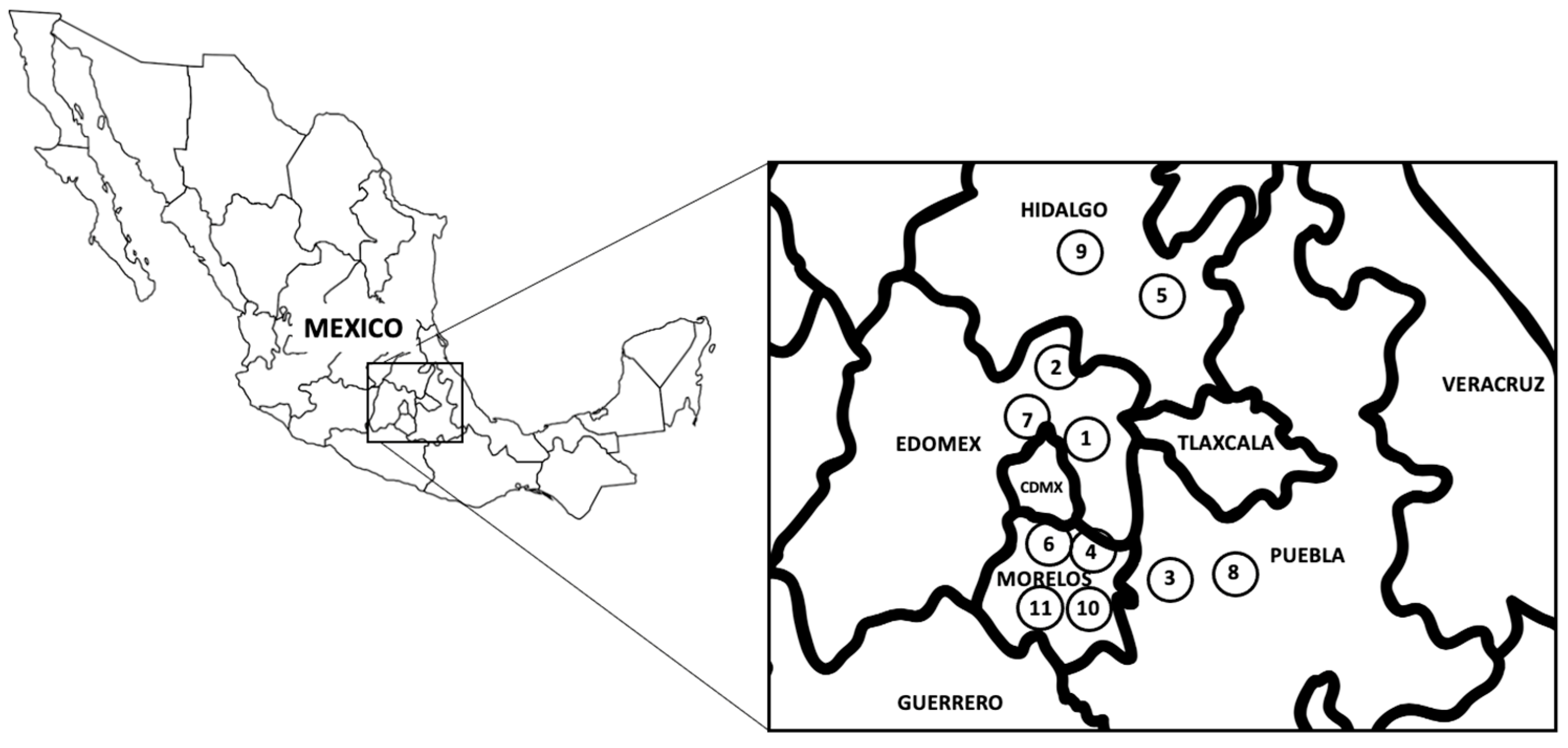
| Sample | Location | Host Species | Parasite Stage | SNP440 | SNP643 | SNP788 | SAAP147 | SAAP215 | SAAP263 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 19.5066° N, 98.8832° W | Bos taurus | ADULT PARASITES | R | A | R | R/K | K | R/K |
| 2 | 19.8128° N, 99.0101° W | Bos taurus | EGGS | G | R | G | R | E/K | R |
| 3 | 18.7649° N, 98.7147° W | Bos taurus | ADULT PARASITES | R | A | G | R/K | K | R |
| 4 | 19.0095° N, 98.9983° W | Ovis aries | EGGS | G | G | G | R | E | R |
| 5 | 20.1437° N, 98.6738° W | Ovis aries | EGGS | G | R | G | R | E/K | R |
| 6 | 18.9848° N, 99.0931° W | Bos taurus | EGGS | G | R | G | R | E/K | R |
| 7 | 19.6861° N, 98.8716° W | Bos taurus | ADULT PARASITES | G | R | G | R | E/K | R |
| 8 | 20.9131° N, 98.4294° W | Ovis aries | EGGS | G | R | G | R | E/K | R |
| 9 | 20.0905° N, 98.3691° W | Ovis aries | EGGS | R | A | G | R/K | K | R |
| 10 | 18.6875° N, 99.1190° W | Bos taurus | EGGS | G | R | G | R | E/K | R |
| 11 | 18.7718° N, 99.3534° W | Bos taurus | ADULT PARASITES | G | R | R | R | E/K | R/K |
| 12 | TCBZ-SUSCEPTIBLE | Ovis aries | ADULT PARASITES | G | G | G | R | E | R |
| 13 | TCBZ-RESISTANT | Ovis aries | ADULT PARASITES | A | A | A | K | K | K |
| TCBZ-Susceptible | TCBZ-Resistant | |
|---|---|---|
| Amino acids in contact with TCBZSOX | F146, R147, A199, S204, C206, G208, C225, T227, H262, R263,G265, L266, P267, P269, H637, Y639 | F146, K147, S204, A199, C206, G208, K210, K212, T227, H262, K263,P264, L266, A281, I282, S283, Y639 |
| TCBZSOX-Binding Pocket Hydrogen Bonds | E198—TCBZSOX C640—TCBZSOX T144—H231 T639—H637 G265—R 263 D201—A199 | D202—TCBZSOX K210—TCBZSOX H262—TCBZSOX P264—TCBZSOX T639—TCBZSOX D202—S204 G208—H262 G208—C206 K210—V217 K263—D623 G265—K263 P264—L266 C225—T227 G623—K212 |
| TCBZSOX-Binding Pocket Ionic Bonds | F146—TCBZSOX | T639—TCBZSOX |
| Binding-Pocket Size Å3 | 1843 | 2233 |
| Vina Score | −5.5 | −8.3 |
| AA Substitutions | 147 R 215 E 263 R | 147 K 215 K 263 K |
| Catalytic site AA Prediction | K210, N211, G214, E215, L216, V217, G256, Y258, L259, Y284, S336, T339, I340. | K210, N211, G214, K215, L216, V217, G256, Y258, L259, Y284, S336, T339, N735. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda-Miranda, E.; Cossío-Bayúgar, R.; Trejo-Castro, L.; Aguilar-Díaz, H. Single Amino Acid Polymorphisms in the Fasciola hepatica Carboxylesterase Type B Gene and Their Potential Role in Anthelmintic Resistance. Pathogens 2023, 12, 1255. https://doi.org/10.3390/pathogens12101255
Miranda-Miranda E, Cossío-Bayúgar R, Trejo-Castro L, Aguilar-Díaz H. Single Amino Acid Polymorphisms in the Fasciola hepatica Carboxylesterase Type B Gene and Their Potential Role in Anthelmintic Resistance. Pathogens. 2023; 12(10):1255. https://doi.org/10.3390/pathogens12101255
Chicago/Turabian StyleMiranda-Miranda, Estefan, Raquel Cossío-Bayúgar, Lauro Trejo-Castro, and Hugo Aguilar-Díaz. 2023. "Single Amino Acid Polymorphisms in the Fasciola hepatica Carboxylesterase Type B Gene and Their Potential Role in Anthelmintic Resistance" Pathogens 12, no. 10: 1255. https://doi.org/10.3390/pathogens12101255
APA StyleMiranda-Miranda, E., Cossío-Bayúgar, R., Trejo-Castro, L., & Aguilar-Díaz, H. (2023). Single Amino Acid Polymorphisms in the Fasciola hepatica Carboxylesterase Type B Gene and Their Potential Role in Anthelmintic Resistance. Pathogens, 12(10), 1255. https://doi.org/10.3390/pathogens12101255