Trypanosoma cruzi STIB980: A TcI Strain for Drug Discovery and Reverse Genetics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Cultivation
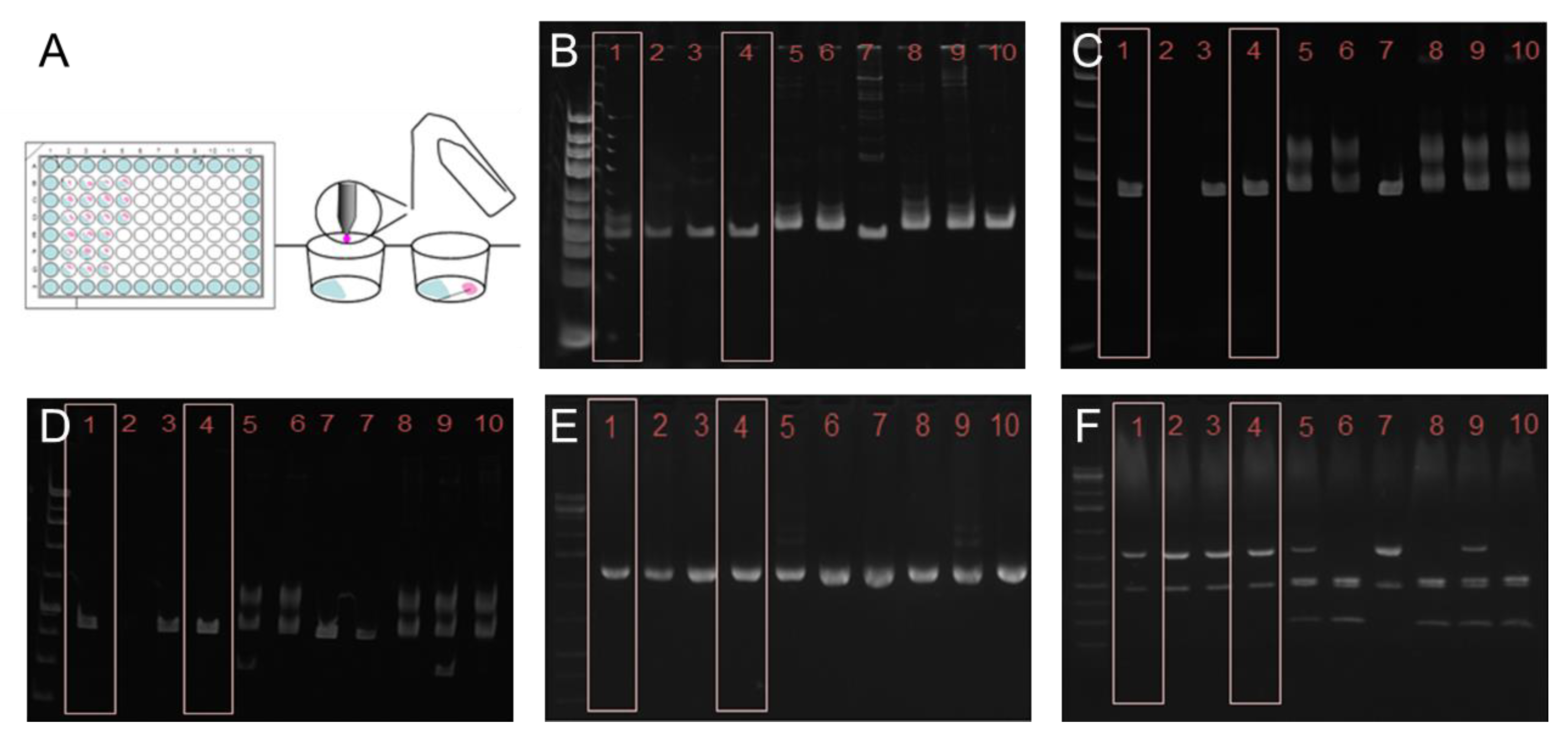
2.2. Cloning of T. cruzi
2.3. Isolation of Genomic DNA
2.4. Genome Sequencing and Assembly
2.5. Optimization of Electroporation
2.6. Generation of Transgenic Lines
2.7. Drug Sensitivity Assay with Epimastigotes
2.8. Flow Cytometry
2.9. High-Content Drug Efficacy Assay
3. Results and Discussion
3.1. Genotyping and Cloning of T. cruzi STIB980
3.2. Genome Sequence of T. cruzi STIB980
3.3. Antibiotic Sensitivity Profile of Epimastigote T. cruzi STIB980
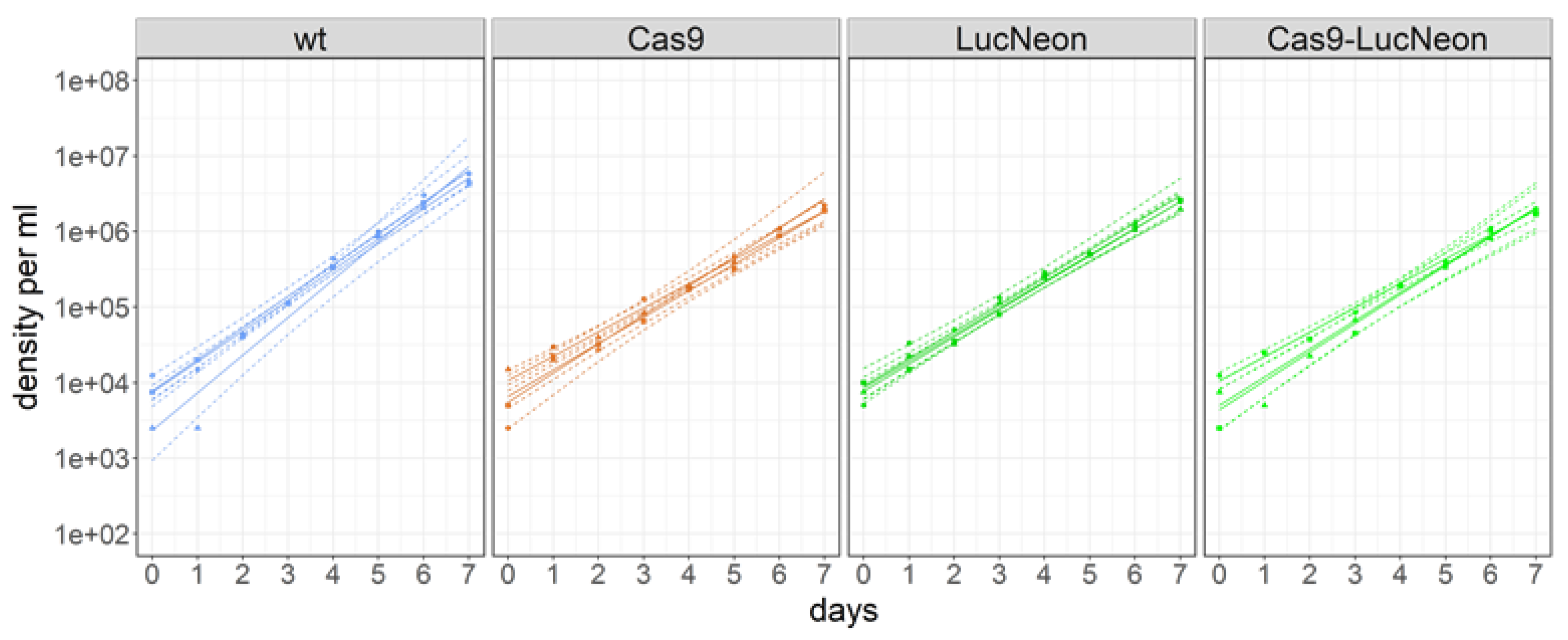
3.4. Optimal Transfection Protocol for T. cruzi STIB980
3.5. Transgenic T. cruzi STIB980 Lines for Drug Testing and Reverse Genetics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef] [PubMed]
- De Fuentes-Vicente, J.A.; Santos-Hernandez, N.G.; Ruiz-Castillejos, C.; Espinoza-Medinilla, E.E.; Flores-Villegas, A.L.; de Alba-Alvarado, M.; Cabrera-Bravo, M.; Moreno-Rodriguez, A.; Vidal-Lopez, D.G. What Do You Need to Know before Studying Chagas Disease? A Beginner’s Guide. Trop. Med. Infect. Dis. 2023, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.M.; Xavier, S.; Roque, A.L.R. Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil. Parasit. Vectors 2018, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Velasquez-Ortiz, N.; Ramirez, J.D. Understanding the oral transmission of Trypanosoma cruzi as a veterinary and medical foodborne zoonosis. Res. Vet. Sci. 2020, 132, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Angheben, A.; Boix, L.; Buonfrate, D.; Gobbi, F.; Bisoffi, Z.; Pupella, S.; Gandini, G.; Aprili, G. Chagas disease and transfusion medicine: A perspective from non-endemic countries. Blood Transfus. 2015, 13, 540–550. [Google Scholar]
- Edwards, M.S.; Montgomery, S.P. Congenital Chagas disease: Progress toward implementation of pregnancy-based screening. Curr. Opin. Infect. Dis. 2021, 34, 538–545. [Google Scholar] [CrossRef]
- Velásquez-Ortiz, N.; Herrera, G.; Hernández, C.; Muñoz, M.; Ramírez, J.D. Discrete typing units of Trypanosoma cruzi: Geographical and biological distribution in the Americas. Sci. Data 2022, 9, 360. [Google Scholar] [CrossRef]
- Wang, W.; Peng, D.; Baptista, R.P.; Li, Y.; Kissinger, J.C.; Tarleton, R.L. Strain-specific genome evolution in Trypanosoma cruzi, the agent of Chagas disease. PLoS Pathog. 2021, 17, e1009254. [Google Scholar] [CrossRef]
- Matsuda, N.M.; Miller, S.M.; Evora, P.R. The chronic gastrointestinal manifestations of Chagas disease. Clinics (Sao Paulo) 2009, 64, 1219–1224. [Google Scholar] [CrossRef]
- Lewis, M.D.; Francisco, A.F.; Taylor, M.C.; Kelly, J.M. A new experimental model for assessing drug efficacy against Trypanosoma cruzi infection based on highly sensitive in vivo imaging. J. Biomol. Screen. 2015, 20, 36–43. [Google Scholar] [CrossRef]
- Ramírez-Toloza, G.; Ferreira, A. Trypanosoma cruzi Evades the Complement System as an Efficient Strategy to Survive in the Mammalian Host: The Specific Roles of Host/Parasite Molecules and Trypanosoma cruzi Calreticulin. Front. Microbiol. 2017, 8, 1667. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Valdez, F.J.; Padilla, A.; Wang, W.; Orr, D.; Tarleton, R.L. Spontaneous dormancy protects Trypanosoma cruzi during extended drug exposure. Elife 2018, 7, e34039. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.I.; Olmo, F.; Atherton, R.L.; Taylor, M.C.; Kelly, J.M. Trypanosoma cruzi amastigotes that persist in the colon during chronic stage murine infections have a reduced replication rate. Open Biol. 2020, 10, 200261. [Google Scholar] [CrossRef] [PubMed]
- Molina, I.; Gomez i Prat, J.; Salvador, F.; Trevino, B.; Sulleiro, E.; Serre, N.; Pou, D.; Roure, S.; Cabezos, J.; Valerio, L.; et al. Randomized trial of posaconazole and benznidazole for chronic Chagas’ disease. N. Engl. J. Med. 2014, 370, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Morillo, C.A.; Waskin, H.; Sosa-Estani, S.; Del Carmen Bangher, M.; Cuneo, C.; Milesi, R.; Mallagray, M.; Apt, W.; Beloscar, J.; Gascon, J.; et al. Benznidazole and Posaconazole in Eliminating Parasites in Asymptomatic T. cruzi Carriers: The STOP-CHAGAS Trial. J. Am. Coll. Cardiol. 2017, 69, 939–947. [Google Scholar] [CrossRef]
- Beilstein, S.; El Phil, R.; Sahraoui, S.S.; Scapozza, L.; Kaiser, M.; Mäser, P. Laboratory Selection of Trypanosomatid Pathogens for Drug Resistance. Pharmaceuticals 2022, 15, 135. [Google Scholar] [CrossRef]
- Taylor, M.C.; Huang, H.; Kelly, J.M. Genetic techniques in Trypanosoma cruzi. Adv. Parasitol. 2011, 75, 231–250. [Google Scholar]
- Docampo, R. Molecular parasitology in the 21st century. Essays Biochem. 2011, 51, 1–13. [Google Scholar] [CrossRef]
- Lander, N.; Chiurillo, M.A. State-of-the-art CRISPR/Cas9 Technology for Genome Editing in Trypanosomatids. J. Eukaryot. Microbiol. 2019, 66, 981–991. [Google Scholar] [CrossRef]
- Kolev, N.G.; Tschudi, C.; Ullu, E. RNA interference in protozoan parasites: Achievements and challenges. Eukaryot. Cell 2011, 10, 1156–1163. [Google Scholar] [CrossRef]
- Fesser, A.F.; Braissant, O.; Olmo, F.; Kelly, J.M.; Mäser, P.; Kaiser, M. Non-invasive monitoring of drug action: A new live in vitro assay design for Chagas’ disease drug discovery. PLoS Negl. Trop. Dis. 2020, 14, e0008487. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.C.; Francisco, A.F.; Jayawardhana, S.; Mann, G.S.; Ward, A.I.; Olmo, F.; Lewis, M.D.; Kelly, J.M. Exploiting Genetically Modified Dual-Reporter Strains to Monitor Experimental Trypanosoma cruzi Infections and Host-Parasite Interactions. Methods Mol. Biol. 2019, 1955, 147–163. [Google Scholar] [PubMed]
- Fernandes, J.F.; Castellani, O. Growth Characteristics and Chemical Composition of Trypanosoma cruzi. Experimental Parasitology 1966, 18, 195–202. [Google Scholar] [CrossRef]
- Mäser, P.; Grether-Bühler, Y.; Kaminsky, R.; Brun, R. An anti-contamination cocktail for the in vitro isolation and cultivation of parasitic protozoa. Parasitol. Res. 2002, 88, 172–174. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data. 2022. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 May 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Otto, T.D. From sequence mapping to genome assemblies. Methods Mol. Biol. 2015, 1201, 19–50. [Google Scholar]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Salzberg, S.L.; Delcher, A.L.; Kasif, S.; White, O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998, 26, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Berná, L.; Rodriguez, M.; Chiribao, M.L.; Parodi-Talice, A.; Pita, S.; Rijo, G.; Alvarez-Valin, F.; Robello, C. Expanding an expanded genome: Long-read sequencing of Trypanosoma cruzi. Microb. Genom. 2018, 4, e000177. [Google Scholar]
- Burkard, G.; Fragoso, C.M.; Roditi, I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007, 153, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.C.; Francisco, A.F.; Jayawardhana, S.; Calderano, S.G.; Lewis, M.D.; Olmo, F.; Beneke, T.; Gluenz, E.; Sunter, J.; Dean, S.; et al. Expanding the toolbox for Trypanosoma cruzi: A parasite line incorporating a bioluminescence-fluorescence dual reporter and streamlined CRISPR/Cas9 functionality for rapid in vivo localisation and phenotyping. PLoS Negl. Trop. Dis. 2018, 12, e0006388. [Google Scholar] [CrossRef] [PubMed]
- Beneke, T.; Madden, R.; Makin, L.; Valli, J.; Sunter, J.; Gluenz, E. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R. Soc. Open Sci. 2017, 4, 170095. [Google Scholar] [CrossRef] [PubMed]
- Olmo, F.; Costa, F.C.; Mann, G.S.; Taylor, M.C.; Kelly, J.M. Optimising genetic transformation of Trypanosoma cruzi using hydroxyurea-induced cell-cycle synchronisation. Mol. Biochem. Parasitol. 2018, 226, 34–36. [Google Scholar] [CrossRef]
- Lewis, M.D.; Fortes Francisco, A.; Taylor, M.C.; Burrell-Saward, H.; McLatchie, A.P.; Miles, M.A.; Kelly, J.M. Bioluminescence imaging of chronic Trypanosoma cruzi infections reveals tissue-specific parasite dynamics and heart disease in the absence of locally persistent infection. Cell Microbiol. 2014, 16, 1285–1300. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Wickham, H. Tidyverse: Easily Install and Load the ‘Tidyverse’. 2017. Available online: https://CRAN.R-project.org/package=tidyverse (accessed on 1 March 2020).
- Wickham, H.; Bryan, J. Readxl: Read Excel Files. 2018. Available online: https://CRAN.R-project.org/package=readxl (accessed on 1 March 2020).
- Messenger, L.A.; Miles, M.A. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015, 151, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B. Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Tropica 2018, 184, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Izeta-Alberdi, A.; Ibarra-Cerdena, C.N.; Moo-Llanes, D.A.; Ramsey, J.M. Geographical, landscape and host associations of Trypanosoma cruzi DTUs and lineages. Parasit. Vectors 2016, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Shanmugasundram, A.; Starns, D.; Bohme, U.; Amos, B.; Wilkinson, P.A.; Harb, O.S.; Warrenfeltz, S.; Kissinger, J.C.; McDowell, M.A.; Roos, D.S.; et al. TriTrypDB: An integrated functional genomics resource for kinetoplastida. PLoS Negl. Trop. Dis. 2023, 17, e0011058. [Google Scholar] [CrossRef]
- Hamilton, P.B.; Lewis, M.D.; Cruickshank, C.; Gaunt, M.W.; Yeo, M.; Llewellyn, M.S.; Valente, S.A.; Maia da Silva, F.; Stevens, J.R.; Miles, M.A.; et al. Identification and lineage genotyping of South American trypanosomes using fluorescent fragment length barcoding. Infect. Genet. Evol. 2011, 11, 44–51. [Google Scholar] [CrossRef]
- Majeau, A.; Murphy, L.; Herrera, C.; Dumonteil, E. Assessing Trypanosoma cruzi Parasite Diversity through Comparative Genomics: Implications for Disease Epidemiology and Diagnostics. Pathogens 2021, 10, 212. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Callejas-Hernández, F.; Rastrojo, A.; Poveda, C.; Gironès, N.; Fresno, M. Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep. 2018, 8, 14631. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Alsford, S.; Kawahara, T.; Glover, L.; Horn, D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol. Biochem. Parasitol. 2005, 144, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Lugo, L.; Díaz-Olmos, Y.; Sáenz-García, J.; Probst, C.M.; DaRocha, W.D. Effective gene delivery to Trypanosoma cruzi epimastigotes through nucleofection. Parasitol. Int. 2017, 66, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Beneke, T.; Gluenz, E. LeishGEdit: A Method for Rapid Gene Knockout and Tagging Using CRISPR-Cas9. Methods Mol. Biol. 2019, 1971, 189–210. [Google Scholar]

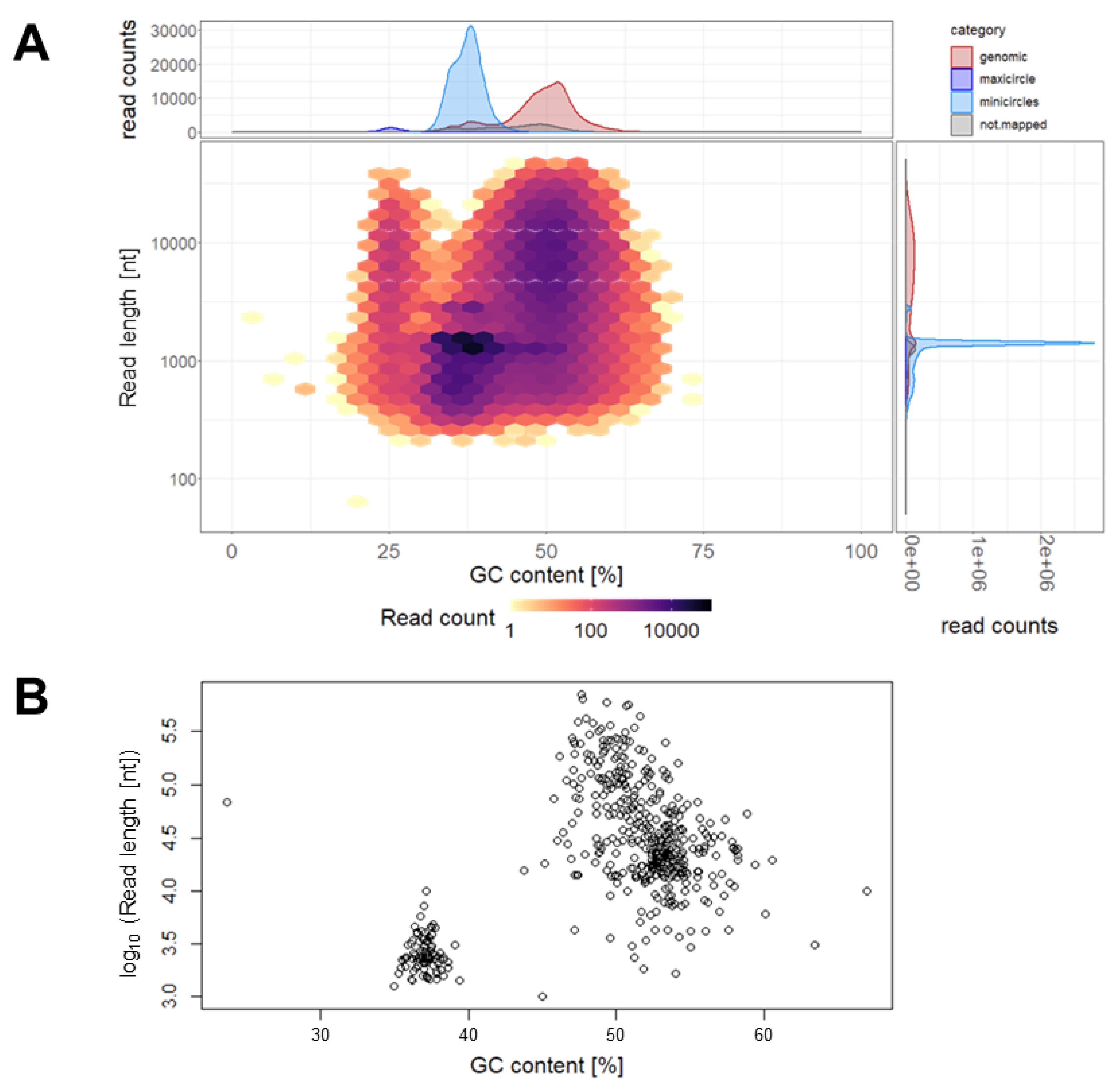

| Total Size (nt) | No. of Contigs | N50 (nt) | Longest Contig (nt) | Shortest Contig (nt) | |
|---|---|---|---|---|---|
| Nuclear | 27,888,483 | 397 | 165,577 | 715,804 | 1660 |
| Minicircles | 248,782 | 91 | 2699 | 10,035 | 1264 |
| Maxicircle | 68,708 | 1 | n.a. | n.a. | n.a. |
| Unknown | 14,070 | 3 | 3090 | 9979 | 1001 |
| Total | 28,220,043 | 492 | 158,042 | 715,804 | 1001 |
| IC50 72 h High Inoculum 1 | IC50 168 h High Inoculum 2 | IC50 168 h Low Inoculum 3 | |
|---|---|---|---|
| Blasticidin | 1.6 (1.2; 2.0) | 0.37 (0.20; 0.54) | 0.32 (0.29; 0.34) |
| Puromycin | 1.3 (1.1; 1.4) | 1.2 (1.1; 1.4) | 0.56 (0.48; 0.64) |
| Hygromycin | 41 (30; 52) | 22 (13; 31) | 37 (28; 46) |
| G418 | 46 (38; 54) | 50 (42; 57) | 31 (25; 37) |
| Phleomycin | 89 (70; 110) | 71 (60; 81) | 27 (23; 32) |
| Benznidazole | 1.9 (0.67; 3.1) | 1.2 (0.90; 1.4) | 0.66 (0.53; 0.79) |
| Nifurtimox | 0.87 (0.58; 1.2) | 0.41 (0.37; 0.46) | 0.24 (0.20; 0.28) |
| DMSO | 3.8 (3.2; 4.5) | 3.2 (−1.1; 7.4) | 1.2 (0.93; 1.4) |
| Program | % Survival | % GFP Expression | Efficiency |
|---|---|---|---|
| X-001 | 55.0 | 6.83 | 0.038 |
| U-033 | 42.5 | 8.15 | 0.035 |
| Z-001 | 32.5 | 7.80 | 0.025 |
| X-014 | 58.8 | 4.16 | 0.024 |
| X-013 | 57.5 | 4.23 | 0.024 |
| X-024 | 65.0 | 3.56 | 0.023 |
| Z-014 | 60.0 | 3.18 | 0.019 |
| X-006 | 57.5 | 2.67 | 0.015 |
| T-020 | 91.3 | 1.11 | 0.010 |
| IC50 (ng/mL) Calculated from Infection Rate | IC50 (ng/mL) Calculated from No. Amastigotes | |||
|---|---|---|---|---|
| wt | LucNeon | wt | LucNeon | |
| Benznidazole | 750 | 840 | 550 | 600 |
| Fexinidazole | 3000 | 4500 | 2900 | 2300 |
| DNDi-6148 | 110 | 110 | 100 | 70 |
| Nifurtimox | 580 | 700 | <410 | 540 |
| Posaconazole | 0.85 | 0.72 | 0.51 | 0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fesser, A.; Beilstein, S.; Kaiser, M.; Schmidt, R.S.; Mäser, P. Trypanosoma cruzi STIB980: A TcI Strain for Drug Discovery and Reverse Genetics. Pathogens 2023, 12, 1217. https://doi.org/10.3390/pathogens12101217
Fesser A, Beilstein S, Kaiser M, Schmidt RS, Mäser P. Trypanosoma cruzi STIB980: A TcI Strain for Drug Discovery and Reverse Genetics. Pathogens. 2023; 12(10):1217. https://doi.org/10.3390/pathogens12101217
Chicago/Turabian StyleFesser, Anna, Sabina Beilstein, Marcel Kaiser, Remo S. Schmidt, and Pascal Mäser. 2023. "Trypanosoma cruzi STIB980: A TcI Strain for Drug Discovery and Reverse Genetics" Pathogens 12, no. 10: 1217. https://doi.org/10.3390/pathogens12101217
APA StyleFesser, A., Beilstein, S., Kaiser, M., Schmidt, R. S., & Mäser, P. (2023). Trypanosoma cruzi STIB980: A TcI Strain for Drug Discovery and Reverse Genetics. Pathogens, 12(10), 1217. https://doi.org/10.3390/pathogens12101217






