Abstract
Understanding the host response to influenza A virus (IAV) infection is vital for developing intervention strategies. The primary barriers for invading respiratory pathogens are the respiratory tract epithelial cells and antimicrobial proteins generated by these cells. The antimicrobial peptide, β-defensin-1, has antiviral activity against both enveloped and non-enveloped viruses. Significant downregulation of β-defensin1 gene (DEFB1) expression was observed when human bronchial epithelial cells (HBEpCs) were exposed to IAV. HBEpCs overexpressing DEFB1 caused a significant reduction in IAV, that was confirmed by IAV matrix gene analysis, plaque assay, and confocal microscopy. DEFB1 expression after transfection with two micro RNAs (miRNAs), hsa-miR-186-5p and hsa-miR-340-5p, provided evidence that DEFB1 expression could be modulated by these miRNAs and hsa-miR-186-5p had a higher binding efficiency with DEFB1. Overexpression of DEFB1 in IAV-infected HBEpCs led to increased NF-κB expression. In a PCR array analysis of 84 transcription factors, either overexpressing DEFB1 or siRNA silencing of DEFB1 expression significantly modulated the expression of signal transducer and activator of transcription 3 (STAT3). In addition, Ingenuity Pathway Analysis (IPA) integrated with PCR array data showed that the JAK1/STAT3 pathway was significantly altered in cells overexpressing DEFB1, suggesting this to be one of the pathways by which defensin regulates IAV replication in HBEpCs. In conclusion, the reduction in IAV copy number in DEFB1 overexpressing cells suggests that β-defensin-1 plays a key role in regulating IAV survival through STAT3 and is a potential target for antiviral drug development.
1. Introduction
The primary barriers for invading pathogens are the airway epithelial cells of the respiratory tract and the antimicrobial peptides produced by these cells. Of these antimicrobials, defensins have a significant role in protecting the cells from pathogens. Defensins are a family of 3–5 kDa cationic peptides rich in cysteine disulfide bonds [1] with a dominant β-sheet structure and three intramolecular disulfide bridges [2] and are part of the innate and adaptive immune system. Defensins are divided into three subfamilies, α, β, and ð defensins, based on their cysteine pairing and molecular structure. Human β-defensin-1 (hBD1) has antiviral activity against both enveloped and non-enveloped viruses [3]. HBD1 is encoded by the gene DEFB1 and produced by different epithelial and bone-marrow-derived cells that have antimicrobial activity against several different types of pathogens, including bacteria, viruses, fungi, and protozoa [1,3,4]. There are over 28 β-defensin genes identified in humans [5]. HBD1 is primarily expressed by epithelial cells at mucosal surfaces, such as those in the gut, skin, airway, mouth, kidney, nose, eyes, mammary glands, and female and male genital tracts [3,6,7]. HBD1 is constitutively produced by epithelial cells and is reported as a potential mediator in mucosal immunity of the lower respiratory tract [8,9]. HBD1 is also expressed in the respiratory epithelium of the lungs and protects the airways against respiratory pathogens [10]. Infections of the respiratory tract by viruses are one of the major causes of morbidity and mortality in humans, and hBD has shown significant antiviral activity against influenza A virus (IAV), respiratory syncytial virus (RSV), and rhinovirus (RV) [11,12]. Antiviral inactivation of these peptides involves multiple mechanisms, such as direct binding of the virus to the peptide and indirect methods of inactivation through the modulation of viral replication, or signaling pathways, and recruitment of immune cells [12].
IAV, which belongs to the Orthomyxoviridae family, is a human respiratory pathogen that causes both seasonal and periodic pandemics [13]. IAV pandemics can be responsible for substantial morbidity and mortality, particularly in high-risk groups. Antiviral response factors present in host tissues possess the ability to reduce viral replication. An increase in inflammatory cytokines is well documented during influenza infections [14]. An RNA interference study showed the dependency of the influenza virus on host factors for replication and survival, and approximately 295 cellular factors were identified as potentially involved in the initial phase of viral multiplication in host cells [15]. Furthermore, several host cell genes are required for IAV survival and replication [16]. IAV induces the production of hBD2 in epithelial cells of the respiratory tract both in vivo and in vitro [7]. In another in vitro study, Madin–Darby canine kidney (MDCK) cells infected with IAV were protected by blocking entry of the virus with recombinant murine β-defensin-2 [17].
MicroRNAs (miRNAs) are small non-coding RNAs that have a significant role in regulating the pathology of infectious viral diseases. MiRNAs are regulators of gene expression, and their increase or decrease in expression alters the down- or upregulation of protein synthesis. Target gene expression is generally repressed through translational inhibition and RNA degradation. Our earlier studies [18,19] have indicated that miRNA profile was significantly changed on exposure to influenza virus in human airway epithelial cells. The miRNA-mediated regulation of hBD1 is still obscure and, hence, studies were conducted to identify the miRNAs regulating the hBD1 synthesis.
As IAV undergoes antigenic shift and drift, vaccines become less effective, and antiviral peptides, such as hBD1, produced by the host are an important line of defense against infection. The function of hBD1 is not well explored in HBEpCs during IAV infection. Here, we provide evidence that microRNAs miR-186-5p and miR-340-5p regulate expression of hBD1. We show that upregulation of hBD1 leads to upregulation of the transcription factor STAT3 and decreased replication of IAV.
2. Materials and Methods
2.1. Propagation of Cells
HBEpCs were obtained from PromoCell (Heidelberg, Germany) and sub-cultured in airway epithelial cell growth media (cat # C-21060) and growth factors following supplier recommendations. Madin–Darby canine kidney (MDCK) cells were cultivated in Eagle’s Minimum Essential Medium (MEM, ATCC), adding 10% fetal bovine serum, penicillin, and streptomycin sulfate (100 µg/mL each). MDCK cells were used for the cultivation of influenza viruses A(H1N1) (A/WSN/33), A(H9N1) A(IWF10), and A(H9N1) (IP10). A(H1N1) (A/WSN/33) was provided by Prof. Robert A. Lamb (Northwestern University, Chicago, IL, USA), and the A(H9N1) viruses were from Daniel Perez (University of Georgia, Athens, GA, USA). Cultivation and maintenance of the viruses were carried out as described [19,20,21].
2.2. Viral Infections
All infections of HBEpCs were performed in 6-well plates at different multiplicity of infection (MOI) as specified for each experiment. Control cells were mock-infected and described as “mock”. Six-well plates were seeded with 1 × 105 cells per well and grown to 80% confluence. The cells were prerinsed with phosphate-buffered saline (PBS) prior to infection experiments, and Modified Hank’s buffer salt solution (MHBSS) was used for diluting the virus to obtain specific MOI. After 45 min of incubation, excess virus was washed off using PBS and then DMEM/F12 media (ATCC) was added with (A(H9N1) strains) or (A(H1N1) (A/WSN/33) without 1.0 µg/mL of TPCK-trypsin (Sigma-Aldrich, St Louis, MO, USA), and were incubated at 35 ± 1 °C and 5% CO2.
2.3. hBD1 Overexpression Studies
HBEpCs were transfected with the open reading frame of DEFB1 cloned in pCMV6-Entry vector (Origene, Rockville, MD, USA) and lipofectamine 2000 (Thermo Fisher Scientific, Carlsbad, CA, USA). Controls were cells transfected with an empty pCMV6 vector. Transfected cells were incubated for 48 h, and then infected with IAV at different MOI. Infected cells were incubated for an additional 24 h. Overexpression of DEFB1 was confirmed by confocal microscopy using anti-hDB1 antibody as described below in Materials and Methods for Microscopy. Cells were harvested at different time points. RNA was isolated and reverse transcriptase polymerase chain reaction was carried out for the matrix copy number (IAV) and the DEFB1 transcripts as described earlier [21].
2.4. Transfection of miRNAs and siRNAs
HBEpC were transfected with miRNA inhibitor oligonucleotide or a mimic oligonucleotide (Thermo Fisher Scientific, Carlsbad, CA, USA) using the lipid-based lipofectamine 2000 reagent diluted in Opti-MEM-I reduced serum medium (Thermo Fisher Scientific) according to the suppliers’ protocol. Briefly, HBEpCs were grown in 6-well plates overnight and the transfection mixture was directly applied to the cells at a final concentration of 25 nM oligonucleotides. Cells were transfected with the same concentrations of scrambled oligonucleotides (Thermo Fisher Scientific) and used as control. The transfected cells were then infected at various MOIs of IAV. Following the incubation, cells were harvested and used for RNA extraction using RNeasy kit (Qiagen, Germantown, MD, USA).
2.5. Defensin β-1 Knock-Down Using siRNAs
The interactions between DEFB1 and transcription factors during influenza A(H1N1) infection were further investigated by silencing DEFB1 expression in HBEpCs with DEFB1.siRNA (Dharmacon, Chicago, IL, USA). Exponentially growing cells were transiently transfected with either 25 nM of DEFB1.siRNA or the scrambled siRNA control using lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours following transfection, cells were harvested and used for RNA extraction using RNeasy kit.
2.6. Plaque Assay for IAV Detection
For viral plaque assay, confluent MDCK cells were propagated in 6-well plates. Confluent monolayers of MDCK cells were prerinsed with PBS and infected with serially diluted cell culture supernatant (800 μL per well) collected from the HBEpC cells exposed to influenza virus. Infected cells were incubated for 45 min at 35 °C. Cells were then washed with PBS and overlayed with 0.6% agarose (Oxoid Ltd., Hampshire, UK) in DMEM/F12 medium, and plates were kept at 35 °C for another 60 h. Following the incubation, cells were treated with 10% formalin and agarose layer was removed. Cells were stained with 1.0% crystal violet and plaque-forming units (PFU) were counted.
2.7. Gene Expression and Transcription Factor PCR Array
Total RNA was isolated from HBEpCs using RNeasy kit (Qiagen) and following the protocol of the supplier. Total RNA (1 µg) was converted to cDNA with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). TaqMan primers for DEFB1, NF-κB, BCL-XL, and glyceraldehyde phosphate dehydrogenase (GAPDH) were purchased from Applied Biosystems (Thermo Fisher Scientific, Foster City, CA, USA). Delta-delta Ct (2−∆∆Ct) method was used for calculating the fold change in gene expression as described earlier [21]. Fold change is the normalized expression in each test sample divided by the normalized expression in the control sample. Fold change values less than one (the control sample) indicate a downregulation of expression and the fold downregulated is the negative inverse of the fold change. Values greater than one (the control sample) indicate the fold upregulation of expression.
PCR array analysis for expression and identification of different transcription factors that are differentially regulated in response to IAV infection was performed using TaqMan™ Array, Human Transcription Factors (Non-Hox), Fast 96-well (Catalogue #4418784, Thermo Fisher Scientific). Gene expression results were normalized to the housekeeping gene HPRT1, and data were analyzed using the supplier’s online software and the 2−∆∆Ct method.
IAV matrix gene expression was quantified using a standard curve with matrix gene-specific primers (Taqman assay) and reported as influenza matrix gene copy number.
2.8. Microscopy
DEFB1 overexpressing HBEpCs were grown on chamber slides (Chamber slide™, Lab-TekII, Thermo Fisher Scientific, Rochester, NY, USA) to reach 80–90% confluency. Subsequently, cells were infected with A(H1N1) for another 18 h. Cells were washed with PBS and treated with 4% formaldehyde (Polysciences Inc., Warrington, PA, USA). Staining of cells was completed with anti-IAV antibodies (MAB8256, Millipore sigma, Billerica, MA, USA), anti-hemagglutinin, anti-influenza PB1, and anti-hBD1 antibodies (Abcam, Waltham, MA, USA) for 1 h, followed by suitable secondary antibodies (Alexa 488 or −555; Thermo Fisher Scientific). Photomicrographs were produced using a Zeiss LSM510 with a 63X oil immersion lens (Carl Zeiss, Obertochen, AG Germany).
2.9. Western Immunoblot Analysis
HBEpC cells were transfected with DEFB1 plasmid or control plasmid for 48 h, cells were then exposed to A(H1N1) for an additional 24 h and protein extracts (30 µg) from the cells were prepared with 30 µL of radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) and analyzed by 10% SDS-PAGE. Separated proteins were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). Membranes were blocked using Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE, USA) and probed with anti-STAT3, anti-phosphorylated-STAT3, and mouse monoclonal anti-GAPDH (Abcam) antibodies. Appropriate IRDye 680 or 800 secondary antibodies (LI-COR Biosciences) were used. Fluorescence detection was performed on the Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE, USA), and the signal intensities of the individual bands were determined. Protein quantitation was carried out using the Image lite software (LI-COR Biosciences).
2.10. Argonaute Immunoprecipitation
Co-immunoprecipitation of Argonaute (Ago) proteins bound to miRNA/mRNA complexes is a powerful tool to validate miRNA targets [22]. Ago Immunoprecipitation (Ago-IP) was completed according to the instructions in the Active Motif kit (Active Motif, Carlsbad, CA, USA). Briefly, cells (HBEpCs) were grown in six-well plates. When cells reached 80% confluency, they were transfected with 25 nM mimics of miR-186-5p and miR-340-5p or negative control (scrambled oligonucleotide of mimic) for 24 h. An isotype control of the antibody alone was run in parallel. Following the manufacturer’s protocol, the Ago-IP precipitated complex was collected, and total RNA was extracted using Trizol reagent (Qiagen). High-Capacity cDNA Reverse Transfection Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) was used for cDNA synthesis. Specific primers for DEFB1 (Taqman primers, Thermo Fisher Scientific) were used for RT-PCR. The data were analyzed by comparing the cells transfected with mimic miRNA or negative control oligonucleotide, and the fold enrichment of DEFB1 mRNA was calculated as described by the manufacturer.
2.11. Ingenuity Pathway Analysis
Data generated from the transcription factor PCR array were used for the Ingenuity Pathway Analysis (IPA) software (IPA, Qiagen, Redwood City, CA, USA) for core mRNA pathway investigation. The software was utilized for the construction of interacting mRNA pathway networks identified within the DEFB1 overexpressing group and were compared to a control group.
2.12. Statistical Analysis
One-way analysis of variance (ANOVA) was used to analyze MOI studies as well as studies of mimic and inhibitor expression, and post hoc pairwise multiple comparisons between means were performed using the Holm–Sidak method with a p-value of <0.05 considered statistically significant using Sigma stat for Windows (Systat Software, Chicago, IL, USA).
3. Results
3.1. Role of Defensin-β1 in IAV Replication
Significant downregulation of DEFB1 expression was observed when HBEpCs were exposed to IAV. The reduction in DEFB1 was observed with both IAV virus strains A(H1N1) and A(H9N1) indicating that the reduction in DEFB1 was not strain-dependent (Figure 1). To further understand the role of DEFB1 during IAV infection, HBEpCs were transfected with either a control plasmid, pCMV6, or pCMV-DEFB1, expression of DEFB1 mRNA was assessed by RT-PCR, and hBD1 protein expression was assessed by confocal microscopy. Both control and DEFB1 overexpressing cells were then infected with A(H1N1) at an MOI of 1 for 24 h. Samples were drawn at different time intervals (Figure 2). Expression of DEFB1 in infected cells remained relatively constant over 24 h (Figure 2A). A(H1N1)-infected cells expressing the control plasmid showed increasing A(H1N1) matrix gene copies over 24 h of infection (1 × 107.5 copies), whereas in DEFB1-overexpressing cells, matrix gene copies were reduced by 99% (1 × 105.5 copies) (Figure 2B). Our results suggest that DEFB1 plays a key role in limiting IAV replication in bronchial epithelial cells. In addition, A(H1N1)-infected cells expressing the control plasmid showed increased viable virus in a plaque assay over 24 h of infection, whereas in DEFB1-overexpressing cells, the viable virus counts were significantly lower (Figure 2C). Our results suggest that DEFB1 plays a key role in limiting IAV replication in bronchial epithelial cells.
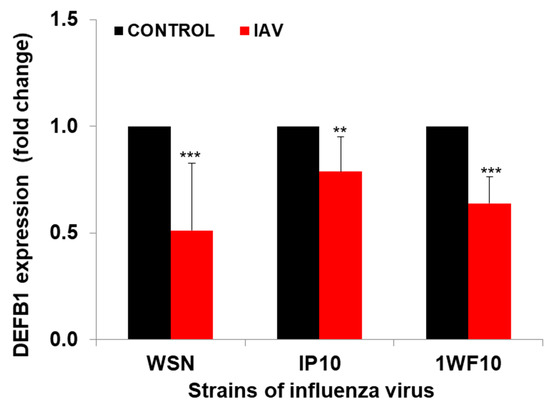
Figure 1.
DEFB1 expression profile. HBEpCs were infected with A(H1N1) and A(H9N1) (1P10 and 1WF10) at MOI of 1.0 for 24 h, and data analyzed by RT-PCR. n = 3. *** = p < 0.001. ** = p < 0.01. Data normalized using the house keeping gene GAPDH.
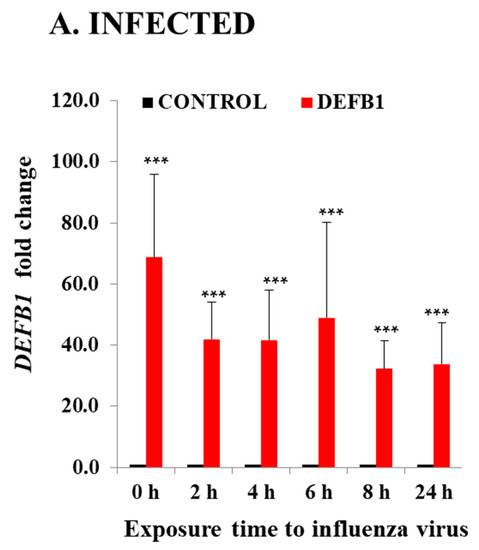
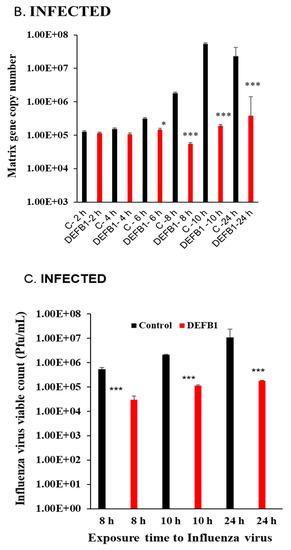
Figure 2.
DEFB1 overexpression protects HBEpCs from A(H1N1) infection. (A) HBEpCs were transfected with DEFB1 plasmid or a control plasmid. Transfected cells were then infected with A(H1N1) at an MOI of 1.0 for 24 h and samples were taken at different time intervals as shown. RNA was extracted and cDNA synthesized and used for RT-PCR analysis. DEFB1 fold change was calculated using the delta-delta Ct method using GAPDH as housekeeping gene. Data presented as ± SEM, *** = p < 0.001, n = 3. (B) Influenza virus matrix gene determined from the RNA extracted from HBEpCs transfected with DEFB1 plasmid or control and infected with A(H1N1). Control cells were transfected with the same plasmid that lacks DEFB1 insert. n = 3 experiments., *** = p < 0.001, * = p < 0.05. (C) Viability of influenza virus in cell culture supernatant quantitated as plaque-forming units (pfu/mL). Data shown as mean ± SE (n = 3 experiments for plaque assay), *** p < 0.001.
Overexpression of hBD1protein in HBEpCs infected with A(H1N1) was next examined using confocal microscopy (Figure 3A). Cells transfected with the DEFB1 expression plasmid showed a marked increase in hBD1 expression (shown as increased red fluorescence). HBEpCs transfected with the control plasmid and subsequently infected with A(H1N1) exhibited an intense green fluorescence (IAV antibody) because of active virus replication. In contrast, cells overexpressing hBD1 showed a sharp reduction in IAV infection (Figure 3A, decreased green fluorescence). Decreased IAV infection in cells overexpressing hBD1 was also indicated by a significant reduction in IAV hemagglutinin protein (Figure 3B). An intense overlapping of red fluorescence was visible in control cells infected with A(H1N1) due to the overexpression of influenza virus PB1 protein (Figure 3C) and a decreased detection of PB1 in cells overexpressing hBD1.
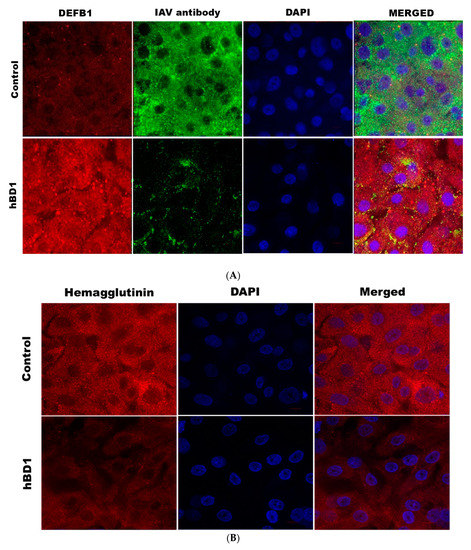
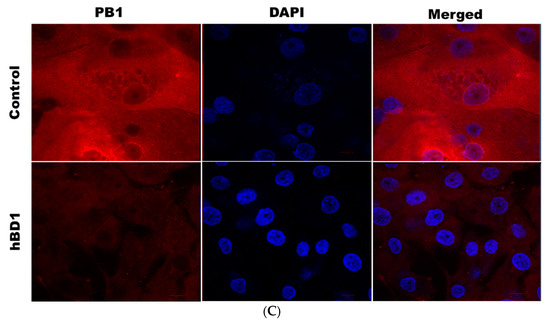
Figure 3.
DEFB1 overexpressing cells show a reduction in different influenza viral proteins. (A) HBEpCs overexpressing the DEFB1 gene or control plasmid were cultured on slides and were infected with A(H1N1) at MOI of 1.0 for 18 h. Cells were stained with anti-hBD1 antibody (red) and IAV antibody MAB8256 (green), and subsequently stained with respective fluorescent secondary antibodies. Nucleus (blue) was stained with DAPI. The microscope settings and contrast adjustments are uniform for a given fluorescence channel across each image. (B) Immunofluorescence of hemagglutinin in DEFB1 overexpressing and control cells. HBEpCs were infected with A(H1N1). DAPI (blue) was used for staining the nucleus of the cells. (C) Immunofluorescence of PB1 expression in cells overexpressing DEFB1 and infected with A(H1N1). Cells were stained DAPI (blue) showing nuclei. All slides were observed using an oil 63X- objective (LSM-510 confocal microscope).
We previously showed that transcription factor NF-κB and its inhibitor NFKBIB regulates IAV replication in HBEpCs [21]. Other studies also have shown that DEFB1 regulates viral replication through NF-κB [12,23]. HBEpCs were infected with A(H1N1) at an MOI of 1.0 and analyzed NF-κB expression in cells overexpressing DEFB1 and control plasmid infected cells (Figure 4). Initially (1–2 h post-infection), levels of NF-κB were significantly higher (p < 0.001) in DEFB1 transfected cells. The highest level of NF-κB expression (1.8-fold) was observed in the early stages of A(H1N1) infection and gradually reduced by 8 h (p < 0.05) and was not significantly different from that of the control cells by 24 h.
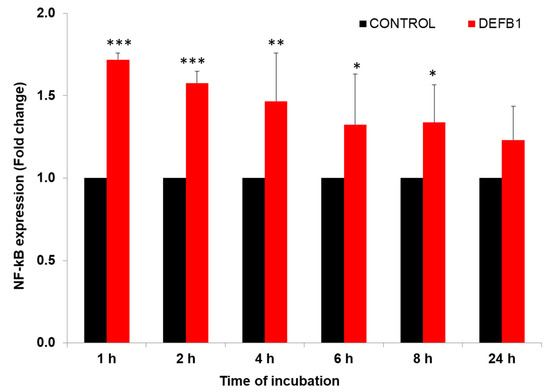
Figure 4.
DEFB1 overexpression modulates the expression of NF-κB. HBEpCs in a 6-well plate were transfected with DEFB1 or a control plasmid lacking the DEFB1 gene and infected with A(H1N1) at MOI of 1 for 24 h. Samples were removed at different time intervals for analysis. RT-PCR conducted using extracted RNA from infected cells. NF-κB fold change expression was analyzed using the delta-delta Ct method. NF-κB values are expressed as ± SEM, *** = p < 0.001; ** = p < 0.01; * = p < 0.05. n = 3. Control represents the cells transfected with plasmid alone and infected with A(H1N1).
3.2. Defensin-β1 Function through STAT3 Pathway
We then used a PCR array consisting of 84 transcription factors that are directly or indirectly involved in viral replication and regulation of the host proteins during IAV infection to search for other transcription factors that may be regulated in response to overexpression of hBD1. Transcription factor expression was first assessed in uninfected HBEpCs that overexpress DEFB1 and in cells transiently transfected with a combination of three DEFB1.siRNAs to downregulate endogenous DEFB1 expression. HBEpCs that overexpress DEFB1 showed more than a two-fold upregulation (more than a two-fold change over the plasmid control) of ATF1, FOXO1, IRF1, RELB, SMAD9, and STAT3, whereas GTF2B, MEF2C, and PPARG were more than two-fold downregulated (negative fold changes compared with plasmid control) (Table 1).

Table 1.
Uninfected HBEpC were transfected with DEFB1 or DEFB1.siRNA showing the expression pattern of transcription factors that were detected. HBEpCs were transfected with DEFB1 and then infected with A(H1N1) showing the expression (fold change) pattern of transcription factors that were detected, along with the house keeping gene HPRT1 (MOI of 1 for 24 h).
In contrast, downregulation of endogenous DEFB1 with siRNAs led to a more than two-fold downregulation of ATF3, CEBPA, ESR1, HIF1A, MEF2C, NFATC2, PPARG, SP3, STAT1, and TFAP2A expression. Interestingly, not all transcription factors that were upregulated in DEFB1 overexpressing cells decreased in cells transfected with DEFB1.siRNA. For example, DEFB1.siRNA transfected cells did not have altered expression of ATF1, RELB, and SMAD9 compared to the control cells.
The JAK/STAT signaling pathway is shown in IPA (Figure 5). Changes in cytokine levels during viral infection have been well documented [18] and the IPA showed that interferon and interleukin attachment to cytokine receptors can induce STAT phosphorylation, which in turn modulates expression of downstream genes, such as C-FOS, IL-6, and BCL-XL, along with cell proliferation. We then verified that one of these downstream target genes, BCL-XL, was upregulated in A(H1N1)-infected HBEpCs that overexpress DEFB1 (Figure 6). A significant upregulation of phosphorylated STAT3 in HBEpCs that overexpress DEFB1 was verified by Western blot analysis (Figure 7A). The cells overexpressing DEFB1 (uninfected) showed significant increase in both STAT3 phosphorylation (Figure 7B) and total STAT3 (Figure 7C) The cells overexpressing DEFB1 infected with 0.5 and 1.0 MOI of A(H1N1) also showed significant changes in both STAT3 phosphorylation (Figure 7B) and total STAT3 (Figure 7C).
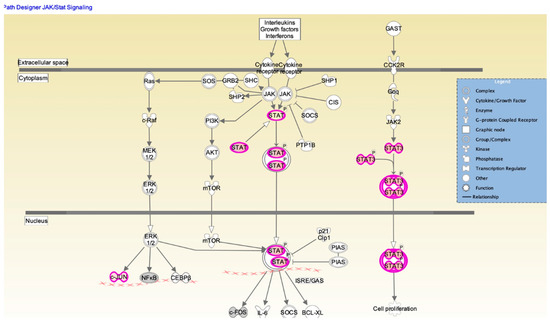
Figure 5.
Ingenuity pathway analysis map depicting the STAT/JAK1 signaling pathway. Signaling molecules showing upregulation are indicated by purple and the intensity of purple represents the signal strength. STAT interactions in cytoplasm and nucleus are shown along with other molecules involved in regulating cell proliferation. Biological network analysis was performed using IPA software. Downstream effector molecules, such as cFOS, IL-6, SOCS, and BCl-XL, are also shown.
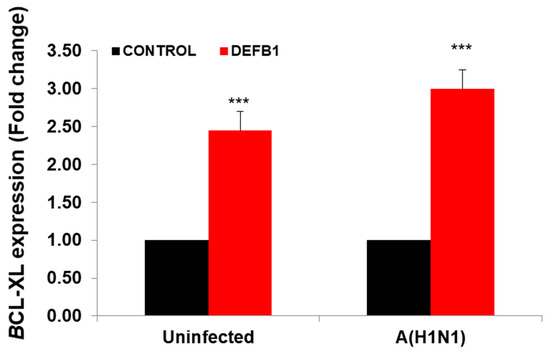
Figure 6.
Expression of BCL-XL showing the downstream pathway of STAT/JAK1 in DEFB1 overexpressing cells. HBEpCs were transfected with DEFB1 or a plasmid control and infected with A(H1N1) at MOI of 1 and incubated for 24 h. Data are expressed as ± SEM, *** = p < 0.001. n = 3. Uninfected control is the cells transfected with plasmid alone and not infected with A(H1N1).
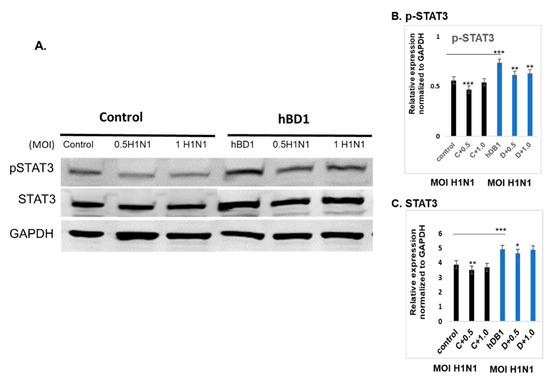
Figure 7.
DEFB1 overexpression modulates STAT3. HBEpCs were transfected with DEFB1 gene containing plasmid or a control vector plasmid. Transfected cells were incubated for 48 h and the cells were infected with 0.5 and 1 MOI of A(H1N1) for an additional 24 h. (A) Western blot of cells overexpressing DEFB1 showing increased phospho-STAT3 protein in both infected and uninfected cells, whereas the control cells infected with A(H1N1) showed a reduction in STAT3 phosphorylation. Housekeeping protein is GAPDH. (B) Quantification of phospho-STAT3, and (C) quantification of STAT3. Quantification was performed using the Odyssey lite software. Data are expressed as ± SEM, *** = p < 0.001; ** = p < 0.01; * = p < 0.05. n = 3 experiments.
3.3. Role of miRNAs Regulating the Expression of Defensin-β1
Our previous studies, along with others, have shown that several miRNAs are differentially regulated during IAV infection of airway epithelial cells [19,21,24]. We therefore assessed whether any of these miRNAs have a role in regulating DEFB1 expression. Using TargetScan (www.targetscan.org; accessed on 15 August 2021) and miRanda database (version 3.3a 2020, accessed 16, August, 2021), we searched for putative miRNAs that target DEFB1. Table 2 shows the top miRNAs that potentially target DEFB1 mRNA. Of these, we selected three miRNAs, miR-340-5p, miR-186-5p, and miR-202-3p, that were differentially regulated in IAV-infected patient sera in our previous study [18]. HBEpCs were transfected separately with miR-340-5p, miR-186-5p, and miR-202-3p inhibitor oligonucleotides (Figure 8A) or miR-340-5p, miR-186-5p, and miR-202-3p mimic oligonucleotides (Figure 8B). The results showed that miR-186-5p mimic and inhibitor could effectively target DEFB1 mRNA. MiR-202-3p did not show any inhibition of DEFB1 with mimic or increase in DEFB1 expression with inhibitor. MiR-340-5p showed a moderate effect on DEFB1 expression in cells transfected with inhibitor and mimic oligonucleotides.

Table 2.
Targeted miRNAs of DEFB1 (data from Target scan).
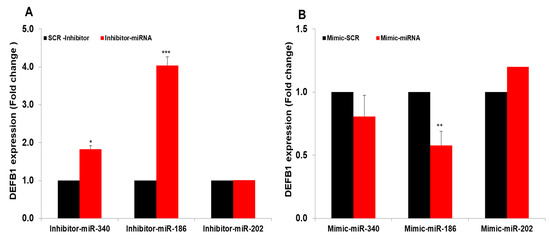
Figure 8.
MiRNAs mimics and inhibitors target DEFB1. (A) HBEpCs were transfected with 25 nM miR-186-5p, miR-340-5p, and miR-202-3p inhibitors or a scrambled oligonucleotide. After 36 h, the DEFB1 transcript level was determined by RT-PCR. Data presented as ± SEM, *** = p < 0.001, and * = p < 0.05. (B) HBEpCs were grown to confluency (80–90%) in a 6-well plate and then transfected for 48 h with 25 nM miR-186-5p, miR-340-5p, and miR-202-3p mimics or a scrambled oligonucleotide. After 36 h of incubation, RNA was isolated and the DEFB1 transcript level was determined by RT-PCR. ** = p < 0.01.
To further confirm the specificity in binding of miR-340 and miR-186 to DEFB1 mRNA, mimics of miR-340-5p, miR-186-5p, or a scrambled oligonucleotide control were used to transfect HBEpCs and then subjected to Argonaute immunoprecipitation (Ago-IP). The results indicate that mimics of miR-186-5p transfected cells showed an 8-fold enrichment of DEFB1 mRNA compared to the scrambled-mimic-transfected cells (Figure 9). In contrast, miR-340-5p mimic led to only a two-fold enrichment of DEFB1 mRNA.
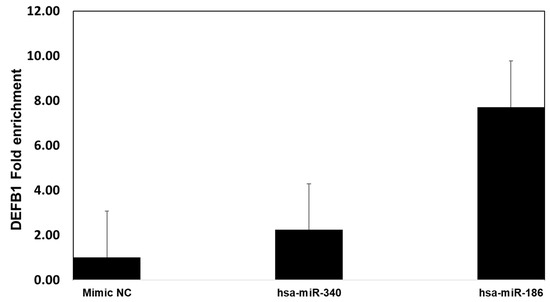
Figure 9.
Confirmation of miRNA target of DEFB1 by Argonaute immunoprecipitation. HBEpCs were transfected with miR-340-5p, miR186-5p mimic, or negative control mimic. Cells were incubated for 24 h. Following the incubation, the cells were lysed, and cell lysate used for immunoprecipitation using the Argonaute 1, 2, 3, or isotype antibody. Fold enrichment of DEFB1 transcript was determined by extracting RNA and running RT-PCR experiment. The fold enrichment was calculated based on the control cells immunoprecipitated using isotype and ago-antibody. n = 2.
4. Discussion
Invading pathogens come in direct contact with airway epithelial cells of the respiratory tract where antimicrobial peptides produced by these cells provide an initial defense mechanism. As microbes evolved, these peptides have retained their antimicrobial properties to the microbial target [25]. Several peptides within the β-defensin family have been investigated for their antimicrobial activity against bacteria and fungi [26]. One peptide, hBD1, encoded by DEFB1, is widely produced by human secretory glands and epithelial cells [27]. In addition, hBD1 is also expressed in plasmacytoid dendritic cells and monocytes and is induced in response to viral challenge [28]. Andresen et al. [29] studied the modulation of hBD1 expression in Chronic Obstructive Pulmonary Disease (COPD) patients and suggested that hBD1 could be used as a biomarker for COPD. Antimicrobial peptides also play a critical role in mammalian innate immunity. Among the viruses, IAV is a seasonal pathogen that undergoes antigenic drifts that can lower the efficacy of an IAV vaccine, emphasizing the importance of an antimicrobial defense mechanism [3]. Human BD1 plays a significant role in protecting airway epithelial cells from invading viruses and bacteria [11]. Our studies showed that DEFB1 gene expression is decreased in cells exposed to IAV over a 24 h time period, and the decrease was consistent with both A(H1N1) and A(H9N1) strains. A significant reduction in IAV infection was observed in HBEpCs overexpressing DEFB1, suggesting that hBD1, could play a major role in regulating IAV infection in airway epithelium. Our study may help determine specifically how hBD1 regulates IAV replication and can lead to a better understanding of the pathophysiology of IAV infections.
In contrast to our results, expression of sheep BD1 (sBD1) mRNA was increased three days post-infection with IAV [30]. In a mouse model, the first six days of IAV A(H3N2) infection showed increased expression of mouse BD1(mBD1) in the trachea and nasosinuses tissues, whereas mBD1 expression in lung tissues increased in the early course of infection and decreased by days 3 and 6 [31]. The difference in expression of defensins reported in in vivo animal studies and our current studies could be attributed to the viability of the virus, specificity of the virus, and the host cells used. hBD1 showed increased expression in other respiratory viruses, such as rhinovirus infections [23]. In vitro studies in HSV-1-infected airway epithelial cells showed an inverse correlation between DEFB1 expression and the virus HSV-1 [27]. Our findings supported this conclusion, as overexpression of DEFB1 significantly reduced IAV copy numbers.
The role of miRNAs in regulating expression of DEFB1 mRNA has not been reported. We showed that DEFB1 expression was significantly altered by two microRNAs, miR-186-5p and miR-340-5p. In previous studies, miR-186-5p and miR-340-5p showed differential expression in IAV-infected airway epithelial cells [18,32]. MiRNAs are regulatory RNAs that control the stability of more than half of mRNAs in humans by recruiting the RNA-induced silencing complex through base-pairing interactions with their targets [33]. In most cases, miRNAs modulate gene expression by binding to complementary segments in the 3′UTR regions of the mRNAs. miRNAs could interact with several conserved mRNA targets that can target different signaling pathways [34,35,36]. Our data indicate that mimics and inhibitors of miR-340-5p and miR-186-5p were able to modify the expression of DEFB1. Generally, each miRNA can be involved in a very complex network by targeting multiple genes [37]. We demonstrated that the mimics of miR-186-5p were able to reduce DEFB1 expression, and that this miRNA may have a role in modulating DEFB1 expression in HBEpCs.
In this study, we also showed that hBD1 can function through the STAT3 pathway. STATs are a family of transcription factors that have vital roles in regulating several biological functions, including immune modulation, angiogenesis, apoptosis, cell differentiation, and induction of inflammatory cytokines [38]. Among the RNA viruses, IAV, human metapneumovirus, and human cytomegalovirus all are able to inhibit phosphorylation of STAT3 [39]. STAT proteins are activated upon phosphorylation and translocate to the nucleus where they activate regulatory genes, cytokines, and growth factors. Our IPA results (Figure 5) support a role for STAT3 in DEFB1 expression and that DEFB1 overexpression may function through the STAT3 pathway. STAT3 is activated at Tyr705, and in this study we used an antibody that was specific to detect Tyr705 phosphorylation of STAT3 (Figure 7, Western blot analysis). In our studies, we showed that overexpression of DEFB1 upregulates STAT3 protein expression and subsequent phosphorylation. Further, our transcription factor array results, which showed increased expression of STAT3 and IRF1 in DEFB1-overexpressing cells, suggests that the DEFB1 may be functional through STAT3 and IRF1, but further studies are needed to confirm this possibility. Combining our previous findings showing that transcription factor NF-κB regulates IAV infection (20), we propose that IAV infection suppresses DEFB1, and by suppressing DEFB1 leads to an alteration of NF-κB and the subsequent IAV matrix gene, and the downregulation of anti-apoptotic proteins BCL-XL and pSTAT3.
5. Conclusions
In conclusion, we found that overexpression of DEFB1, the gene encoding hBD1, significantly reduced IAV matrix gene copies in human airway epithelial cells. Two miRNAs, miR-186-5p and miR-340-5p, regulate expression of DEFB1. The role of STAT3 was further visualized by IPA analysis where the STAT pathway was significantly altered by overexpression of DEFB1. The reduction in viable IAV in DEFB1 overexpressing cells suggests that hBD1 plays an important role in regulating IAV survival through the STAT3 pathway and is a potential target to develop therapies against IAV infections.
Author Contributions
Conceptualization, S.O. and J.D.N.; methodology, S.O.; software, S.O.; validation, S.O. and J.D.N.; formal analysis, S.O.; investigation, S.O.; resources, J.D.N.; data curation, S.O.; writing—original draft preparation, S.O.; writing—review and editing, S.O. and J.D.N.; visualization, S.O.; supervision, J.D.N.; project administration, J.D.N.; funding acquisition, J.D.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ARNT | Aryl hydrocarbon receptor nuclear translocator |
| ATF1 | Activating transcription factor 1 |
| ATF2 | Activating transcription factor 2 |
| ATF3 | Activating transcription factor 3 |
| ATF4 | Activating transcription factor 4 |
| CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta |
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma |
| CREB1 | CAMP responsive element binding protein 1 |
| CREBBP | CREB binding protein |
| CTNNB1 | Catenin (cadherin-associated protein), beta 1 |
| DR1 | Downregulator of transcription 1, TBP-binding |
| E2F1 | E2F transcription factor 1 |
| E2F6 | E2F transcription factor 6 |
| EGR1 | Early growth response 1 |
| ELK1 | ELK1, member of ETS oncogene family |
| ESR1 | Estrogen receptor 1 |
| ETS1 | V-Ets erythroblastosis virus E26 oncogene homolog 1 |
| ETS2 | V-Ets erythroblastosis virus E26 oncogene homolog 2 |
| FOS | Fos Proto-Oncogene, AP-1 Transcription Factor Subunit |
| FOXA2 | Forkhead box A2 |
| FOXO1 | Forkhead box O1 |
| GATA1 | GATA binding protein 1 (globin transcription factor 1) |
| GATA2 | GATA binding protein 2 |
| GATA3 | GATA binding protein 3 |
| GTF2B | General transcription factor IIB |
| GTF2F1 | General transcription factor IIF, polypeptide 1, 74 kDa |
| HAND1 | Heart and neural crest derivatives expressed 1 |
| HAND2 | Heart and neural crest derivatives expressed 2 |
| HDAC1 | Histone deacetylase 1 |
| HIF1A | Hypoxia inducible factor 1, alpha subunit |
| HNF4A | Hepatocyte nuclear factor 4, alpha |
| HOXA5 | Homeobox A5 |
| HSF1 | Heat shock transcription factor 1 |
| ID1 | Inhibitor of DNA binding 1 |
| IRF1 | Interferon regulatory factor 1 |
| JUN | Jun proto-oncogene |
| JUNB | Jun B proto-oncogene |
| MAX | MYC associated factor X |
| MEF2A | Myocyte enhancer factor 2A |
| MEF2B | Myocyte enhancer factor 2B |
| MEF2C | Myocyte enhancer factor 2C |
| NFAT5 | Nuclear factor of activated T-cells 5, tonicity-responsive |
| NFATC1 | Nuclear factor of activated T-cells, calcineurin-dependent 1 |
| NFATC2 | Nuclear factor of activated T-cells, calcineurin-dependent 2 |
| NFATC3 | Nuclear factor of activated T-cells, calcineurin-dependent 3 |
| NFATC4 | Nuclear factor of activated T-cells, calcineurin-dependent 4 |
| NFKB1 | Nuclear factor of kappa B 1 |
| NFYB | Nuclear transcription factor Y, beta |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 |
| PPARA | Peroxisome proliferator-activated receptor alpha |
| PPARG | Peroxisome proliferator-activated receptor gamma |
| RB1 | Retinoblastoma 1 |
| REL | REL V-rel reticuloendotheliosis viral oncogene homolog |
| RELA | V-rel reticuloendotheliosis viral oncogene homolog A |
| RELB | RELB V-rel reticuloendotheliosis viral oncogene homolog B |
| SMAD1 | SMAD family member 1 |
| SMAD4 | SMAD family member 4 |
| SMAD5 | SMAD family member 5 |
| SMAD9 | SMAD family member 9 |
| SP1 | Sp1 transcription factor |
| SP3 | Sp3 transcription factor |
| STAT1 | Signal transducer and activator of transcription 1, |
| STAT2 | Signal transducer and activator of transcription 2, |
| STAT3 | Signal transducer and activator of transcription 3 |
| STAT4 | Signal transducer and activator of transcription 4 |
| STAT5A | Signal transducer and activator of transcription 5A |
| STAT5B | Signal transducer and activator of transcription 5B |
| STAT6 | Signal transducer and activator of transcription 6 |
| TBP | TATA box binding protein |
| TCF7L2 | Transcription Factor 7 Like 2 |
| TFAP2A | Transcription factor AP-2 alpha |
| TGIF1 | TGFB-induced factor homeobox 1 |
| TP53 | Tumor protein p53 |
References
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Becknell, B.; Spencer, J.D.; Carpenter, A.R.; Chen, X.; Singh, A.; Ploeger, S.; Kline, J.; Ellsworth, P.; Li, B.; Proksch, E.; et al. Expression and antimicrobial function of beta-defensin 1 in the lower urinary tract. PLoS ONE 2013, 8, e77714. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef]
- Schutte, B.C.; Mitros, J.P.; Bartlett, J.A.; Walters, J.D.; Jia, H.P.; Welsh, M.J.; Casavant, T.L.; McCray, P.B., Jr. Discovery of five conserved beta -defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 2002, 99, 2129–2133. [Google Scholar] [CrossRef] [PubMed]
- Garreis, F.; Schlorf, T.; Worlitzsch, D.; Steven, P.; Bräuer, L.; Jäger, K.; Paulsen, F.P. Roles of human β-defensins in innate immune defense at the ocular surface: Arming and alarming corneal and conjunctival epithelial cells. Histochem. Cell Biol. 2010, 134, 59–73. [Google Scholar] [CrossRef]
- Doss, M.; White, M.R.; Tecle, T.; Hartshorn, K.L. Human defensins and LL-37 in mucosal immunity. J. Leukoc. Biol. 2010, 87, 79–92. [Google Scholar] [CrossRef]
- Ryan, L.K.; Diamond, G. Modulation of Human β-Defensin-1 Production by Viruses. Viruses 2017, 9, 153. [Google Scholar] [CrossRef]
- Wilson, S.S.; Wiens, M.E.; Holly, M.K.; Smith, J.G. Defensins at the Mucosal Surface: Latest Insights into Defensin-Virus Interactions. J. Virol. 2016, 90, 5216–5218. [Google Scholar] [CrossRef]
- Diamond, G.; Beckloff, N.; Ryan, L.K. Host defense peptides in the oral cavity and the lung: Similarities and differences. J. Dent. Res. 2008, 87, 915–927. [Google Scholar] [CrossRef]
- Findlay, E.G.; Currie, S.M.; Davidson, D.J. Cationic host defence peptides: Potential as antiviral therapeutics. BioDrugs 2013, 27, 479–493. [Google Scholar] [CrossRef]
- Ding, J.; Chou, Y.Y.; Chang, T.L. Defensins in viral infections. J. Innate Immun. 2009, 1, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008, 3, 499–522. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Doyle, W.; Whiteside, T.; Fireman, P.; Hayden, F.G.; Skoner, D. Increased interleukin-6 levels in nasal lavage samples following experimental influenza A virus infection. Clin. Diagn. Lab. Immunol. 1998, 5, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; et al. Human host factors required for influenza virus replication. Nature 2010, 463, 813–817. [Google Scholar] [CrossRef]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef]
- Gong, T.; Jiang, Y.; Wang, Y.; Yang, D.; Li, W.; Zhang, Q.; Feng, W.; Wang, B.; Jiang, Z.; Li, M. Recombinant mouse beta-defensin 2 inhibits infection by influenza A virus by blocking its entry. Arch. Virol. 2010, 155, 491–498. [Google Scholar] [CrossRef]
- Othumpangat, S.; Lindsley, W.G.; Beezhold, D.H.; Kashon, M.L.; Burrell, C.N.; Mubareka, S.; Noti, J.D. Differential Expression of Serum Exosome microRNAs and Cytokines in Influenza A and B Patients Collected in the 2016 and 2017 Influenza Seasons. Pathogens 2021, 10, 149. [Google Scholar] [CrossRef]
- Othumpangat, S.; Noti, J.D.; Blachere, F.M.; Beezhold, D.H. Expression of non-structural-1A binding protein in lung epithelial cells is modulated by miRNA-548an on exposure to influenza A virus. Virology 2013, 447, 84–94. [Google Scholar] [CrossRef]
- Othumpangat, S.; Noti, J.D.; Beezhold, D.H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014, 468–470, 256–264. [Google Scholar] [CrossRef]
- Othumpangat, S.; Bryan, N.B.; Beezhold, D.H.; Noti, J.D. Upregulation of miRNA-4776 in Influenza Virus Infected Bronchial Epithelial Cells Is Associated with Downregulation of NFKBIB and Increased Viral Survival. Viruses 2017, 9, 94. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V., Jr.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods 2008, 44, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Dauletbaev, N.; Gropp, R.; Frye, M.; Loitsch, S.; Wagner, T.O.; Bargon, J. Expression of human beta defensin (HBD-1 and HBD-2) mRNA in nasal epithelia of adult cystic fibrosis patients, healthy individuals, and individuals with acute cold. Respiration 2002, 69, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Lai, W.; Qin, Y.; Zhang, T.; Wang, S.; Ye, X. MicroRNA-33a disturbs influenza A virus replication by targeting ARCN1 and inhibiting viral ribonucleoprotein activity. J. Gen. Virol. 2016, 97, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Sahl, H.-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Krishnakumari, V.; Rangaraj, N.; Nagaraj, R. Antifungal Activities of Human Beta-Defensins HBD-1 to HBD-3 and Their C-Terminal Analogs Phd1 to Phd3. Antimicrob. Agents Chemother. 2009, 53, 256–260. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, I.; Lehrer, R.I. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996, 396, 319–322. [Google Scholar] [CrossRef]
- Ryan, L.K.; Dai, J.; Yin, Z.; Megjugorac, N.; Uhlhorn, V.; Yim, S.; Schwartz, K.D.; Abrahams, J.M.; Diamond, G.; Fitzgerald-Bocarsly, P. Modulation of human beta-defensin-1 (hBD-1) in plasmacytoid dendritic cells (PDC), monocytes, and epithelial cells by influenza virus, Herpes simplex virus, and Sendai virus and its possible role in innate immunity. J. Leukoc. Biol. 2011, 90, 343–356. [Google Scholar] [CrossRef]
- Andresen, E.; Gunther, G.; Bullwinkel, J.; Lange, C.; Heine, H. Increased expression of beta-defensin 1 (DEFB1) in chronic obstructive pulmonary disease. PLoS ONE 2011, 6, e21898. [Google Scholar] [CrossRef]
- Grubor, B.; Gallup, J.M.; Meyerholz, D.K.; Crouch, E.C.; Evans, R.B.; Brogden, K.A.; Lehmkuhl, H.D.; Ackermann, M.R. Enhanced surfactant protein and defensin mRNA levels and reduced viral replication during parainfluenza virus type 3 pneumonia in neonatal lambs. Clin. Diagn. Lab. Immunol. 2004, 11, 599–607. [Google Scholar] [CrossRef]
- Chong, K.T.; Thangavel, R.R.; Tang, X. Enhanced expression of murine beta-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology 2008, 380, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Khongnomnan, K.; Makkoch, J.; Poomipak, W.; Poovorawan, Y.; Payungporn, S. Human miR-3145 inhibits influenza A viruses replication by targeting and silencing viral PB1 gene. Exp. Biol. Med. 2015, 240, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Kaeuferle, T.; Bartel, S.; Dehmel, S.; Krauss-Etschmann, S. MicroRNA methodology: Advances in miRNA technologies. Methods Mol. Biol. 2014, 1169, 121–130. [Google Scholar] [PubMed]
- Izaurralde, E. Breakers and blockers-miRNAs at work. Science 2015, 349, 380–382. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.; Lee, S.; Lee, B.; Kim-Ha, J. Phylogenetic comparison of oskar mRNA localization signals. Biochem. Biophys. Res. Commun. 2014, 444, 98–103. [Google Scholar] [CrossRef] [PubMed]
- De Tullio, G.; De Fazio, V.; Sgherza, N.; Minoia, C.; Serrati, S.; Merchionne, F.; Loseto, G.; Iacobazzi, A.; Rana, A.; Petrillo, P.; et al. Challenges and opportunities of microRNAs in lymphomas. Molecules 2014, 19, 14723–14781. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.A.; Zhong, Z.; Wen, Z.; Darnell, J.E., Jr. STAT signaling is active during early mammalian development. Dev. Dyn. 1997, 208, 190–198. [Google Scholar] [CrossRef]
- Kuchipudi, S.V. The Complex Role of STAT3 in Viral Infections. J. Immunol. Res. 2015, 2015, 272359. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).