Abstract
Mycoplasma gallisepticum (MG) can cause respiratory disease in chickens and result in serious economic losses in the chicken industry. The use of live vaccines has been a favorable option for the control of MG infection in multi-age commercial layers and broiler breeders. There are three live vaccines, including ts-11, 6/85, and F strain, that have been commonly used in various parts of the world, including South Korea. The definitive diagnosis of the infection, therefore, requires the differentiation of wild-type field strains of MG from the vaccine strains used. Thus, we aimed to develop a novel multiplex PCR assay to discriminate between vaccine strains (ts-11, 6/85, and F strain) and wild-type field strains of MG isolated from infected chickens. We designed four novel primer sets that are each specific to MG species, ts-11, 6/85, and F strain. The multiplex PCR assay using the primer sets differentially identified wild-type and vaccine strains of MG but did not detect other avian bacteria. The detection limit of this assay was 250 fg/μL of genomic DNA of each strain tested. In addition, this assay was applied to 36 MG strains isolated from chickens over the past 20 years in South Korea. As a result, the assay identified 22 wild-type strains and 14 vaccine strains. Consequently, the novel multiplex PCR assay can discriminate between vaccine and wild-type field strains of MG and could be a valuable tool for the diagnosis of MG infection in MG-vaccinated chicken flocks.
Keywords:
Mycoplasma gallisepticum; chicken; live vaccine; multiplex PCR; diagnosis; differentiation 1. Introduction
Mycoplasma gallisepticum (MG) is an avian pathogen that phylogenetically belongs to the class Mollicutes. This pathogen is known to cause chronic respiratory disease involving airsacculitis in chickens. Symptoms of MG infection include nasal discharge, sneezing, coughing, and conjunctivitis [1,2,3]. MG infection is also transmitted vertically through infected eggs and horizontally by direct or indirect contact with infected birds. It is also a worldwide problem, as it reduces egg production, hatchability, and feed efficiency on poultry farms, causing huge economic losses in the poultry industry in various parts of the world [4]. In addition, this infection may be exacerbated by complications with E. coli and other viral pathogens, such as infectious bronchitis virus, rather than a single infection [1]. Utilizing live vaccines has been a favorable option for the control of MG infection in multi-age commercial layers and broiler breeders in many countries, including South Korea [2,5,6,7]. Three live vaccines, including ts-11, 6/85, and F strain, are commercially available for preventing MG infection worldwide.
Diagnostic methods for MG infection include serological tests, such as serum plate agglutination test (SPA) or enzyme-linked immunosorbent assay (ELISA), bacterial cultures, and genetic methods using polymerase chain reaction (PCR) and real-time PCR [3,8,9,10,11]. However, the definitive diagnosis of the infection requires the differential identification of wild-type field strains of MG and vaccine strains in those areas where live MG vaccines are used. Currently, MG strains can be distinguished by the genotyping offered by gene-targeted sequencing (GTS) using four primers and multilocus sequence typing (MLST) using six housekeeping genes, allowing for the differentiation between wild-type and vaccine strains of MG [12,13,14]. However, this approach takes a very long time because MG strains are differentiated through PCR and subsequent sequence analysis.
Therefore, the purpose of this study was to develop a fast and accurate multiplex PCR assay that can differentiate wild-type MG strains from the three types of MG vaccine strains (ts-11, 6/85, and F strain) in a single reaction.
2. Materials and Methods
2.1. Bacterial Strains and DNA Extraction
This study included five reference strains (including three vaccine strains) and thirty-six field strains of MG, including twenty-two wild-type strains and fourteen vaccine strains (one ts-11, one 6/85, and twelve F strains), along with twenty-seven strains of other bacterial species (Table 1). The three vaccine strains, ts-11 (Vaxsafe® MG, Boehringer Ingelheim., Seoul, South Korea), 6/85 (Nobilis® MG 6/85, MSD Animal Health, USA), and F strain (PoulShot® MG-F, CAVAC Inc., Daejeon, South Korea), were purchased from vaccine companies. MG field strains were isolated from the tracheas (tissues and cotton swabs) from chickens from different commercial poultry flocks during the period of 2005–2021. These isolates were confirmed by using PCR as reported in the WOAH standard diagnostic manual [3] and were also differentiated into wild-type strains and vaccine strains using GTS analysis [12].

Table 1.
Bacterial strains used to test the sensitivity and specificity of the multiplex PCR assay.
DNA extraction from cultured strains was performed according to the manufacturer’s instruction using a DNA blood and tissue kit (Qiagen Korea Ltd., Seoul, South Korea), and the DNA was stored at −20 °C.
2.2. Sequence Analysis and Primer Design
To identify genetic regions specific to MG species and the three vaccine strains (ts-11, 6/85, and F strain), the whole-genome sequences of eighteen MG strains (GenBank accession nos. NZ_CP044225, NZ_CP044224, NC_017503, NZ_CP028146, NZ_ CP028147, NZ_CP044226, NC_023030, NC_004829, NC_017502, NC_018406, NC_018407, NC_018408, NC_018409, NC_018410, NC_018411, NC_018412, NC_018413, and NZ_LS991952) were downloaded from the NCBI genome database and analyzed using CLC GenomeWorkbench (ver. 20) software (Qiagen, Aarhus, Denmark). Candidate regions specific to each of the three MG vaccine strains and MG species were validated with a homology search using BLAST on the NCBI website. Four primer sets that are each specific to MG species, specifically strain ATCC 15302, as well as ts-11, 6/85, and F strain, were designed using CLC Main Workbench 20. These primer sequences and the sizes of the amplification products are listed in Table 2.

Table 2.
Primer sets used for multiplex PCR analysis for the differential identification of MG strains.
2.3. Multiplex PCR Assay
A multiplex PCR assay was performed to discriminate between the three MG vaccine strains and the field strains using four target regions (parE, RS03710-rlmB, scpA, and MGF_RS03965). PCR amplification was optimized using AccuPower® Multiplex PCR Premix (Bioneer, Daejeon, South Korea) (Table 3). The amplified products were detected under UV light after the electrophoresis of a 1.5% agarose gel containing 5 μL of RedSafe staining solution (iNtRON, Seongnam, South Korea) and 100 ml of TAE buffer. The product sizes of the specific regions were determined by using a 100 bp DNA ladder (Bioneer) as a size marker.

Table 3.
The optimal amplification conditions for discriminating between field strains and vaccine strains of MG using multiplex PCR.
2.4. Specificity and Sensitivity of the Multiplex PCR Assay
Bacteria used to assess the specificity of the multiplex PCR assay were determined using DNA from 27 non-MG strains listed in Table 1. The amplification of the positive control was conducted by mixing DNA templates from each of the four strains (ATCC 15302, ts-11, 6/85, and F strain). The sensitivity of the multiplex PCR assay was evaluated using 5 MG reference strains (ATCC 15302, ts-11, 6/85, F strain, and R-low), 22 MG wild-type strains, and 14 MG field vaccine strains identified by a previously reported GTS assay.
2.5. Limit of Detection of the Multiplex PCR Assay
The detection limit of the multiplex PCR assay was determined using 10-fold serial dilutions of a mixture (2.5 ng–2.5 fg/each strain) of 10 ng of the DNA templates from each of four strains (MG ATCC 15302, ts-11, 6/85, and F strain) in a single tube. DNA concentrations were measured using a Nanodrop ND-1000 UV/UVS spectrophotometer (Nanodrop Tech., Wilmington, DE, USA).
2.6. Evaluation Using Clinical Samples
The multiplex PCR assay was validated with tracheal tissues and swabs from chickens from four poultry farms. Two broiler breeder farms were unvaccinated, and trachea samples were collected at 61 wks and 66 wks of age. Laying hen and broiler breeder farms were vaccinated with ts-11 and F strain, and samples were collected at 28 wks and 38 wks of age.
3. Results
3.1. Primer Design
Compared to the 18 strains of MG, a specific region of the ts-11 vaccine strain was confirmed in the interspace section (positions 879439–879607) located from RS03710 to the rlmB region. This section showed a similarity of 79–84% with other MG strains, and primer sets were designed for this site. For the 6/85 vaccine strain, a specific site was identified at positions 525779 to 526349 of the scpA region. This site showed a similarity of 91% with other MG strains. BLAST analysis of the MGF_RS03965 (positions 598259–598621) region of the F vaccine strain showed that it had 100% similarity with all F strain series (F strain, f99 Avipro, and f99 lab) (Table 2 and Figure 1). In addition, a 245 bp section of the parE region of MG species specifically detected MG.


Figure 1.
Sequence alignments of four regions specific to each of the four different strains (MG 5302, ts-11, 6/85, and F strain). (A) A sequence alignment for MG common gene. (B) Specific region of ts-11. (C) Specific region of 6/85. (D) Specific region of F strain was not present in other MG strains.
3.2. Specificity and Sensitivity of the Multiplex PCR Assay
To determine the specificity of the multiplex PCR assay, we investigated 5 MG strains (ATCC 15302, ts-11, 6/85, F strain, and R-low) and 27 different avian pathogens, as listed in Table 1. As a result, the four primer sets generated specific fragments of 245, 169, 571, and 333 bp for MG species, ts-11, 6/85, and F strain, respectively (Figure 2).
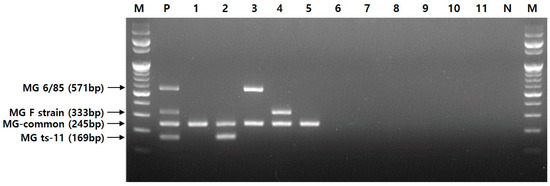
Figure 2.
MG species and representative bacteria tested using a novel multiplex PCR assay. Lane M, 100 bp DNA ladder; lane P, positive; lane 1, MG ATCC 15302; lane 2, MG ts-11 (vaccine); lane 3, MG 6/85 (vaccine); lane 4, MG F strain (vaccine); lane 5, MG R-low; lane 6, M. synoviae; lane 7, M. hyopneumoniae; lane 8, M. hyorhinis; lane 9, A. avium; lane 10, E. coli ATCC 25922; lane 11, P. canis; lane N, negative.
However, 27 non-MG strains were negative in multiplex PCR assay (Table 1). We also used 36 isolates to confirm that the sensitivity of the multiplex PCR method was suitable. MG isolates were classified into 22 field strains and 14 vaccine strains. The fourteen vaccine strains distinctly differentiated one ts-11 strain (no. 40), one 6/85 strain (no. 63), and twelve F strains (nos. 42, 52–55, 57, 62, and 64–68) (Table 1 and Figure 3).
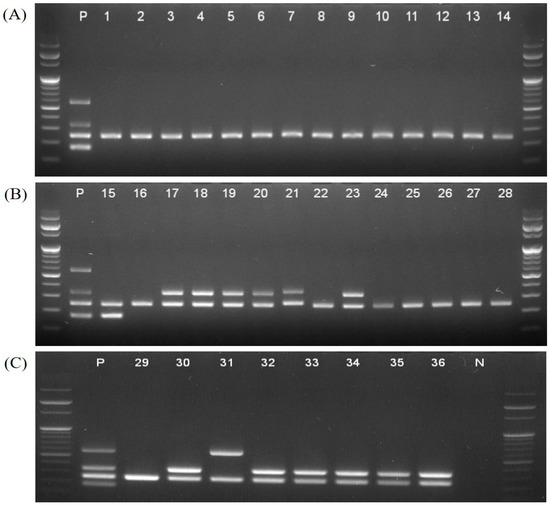
Figure 3.
Amplicons of 36 isolates tested using the multiplex PCR assay. (A) Wild-type strains (nos. 1–14), (B) one ts-11 strain (no.15), six F strains (nos. 17–21, and 23) and seven wild-type strains (nos. 16, 22 and 24–28), and (C) one wild-type strain (no. 29), one 6/85 strain (no.31) and six F strains (nos. 30 and 32–36).
3.3. Detection Limit of the Multiplex PCR Assay
The detection limit of the multiplex PCR assay was determined using 10-fold serial dilutions of a mixture of genomic DNA concentrations of the four MG strains (ATCC 15302, ts-11, 6/85, and F strain), ranging from 2.5 ng to 2.5 fg. The assay was able to detect up to 250 fg/μl (Figure 4).
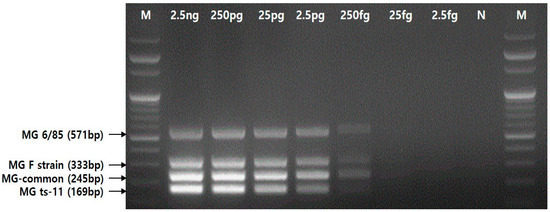
Figure 4.
The detection limit of the multiplex PCR assay using the four primer sets.
3.4. Efficiency on Clinical Samples
To assess the application possibilities of the four primer sets on clinical samples, DNA extracted from the tracheal tissues and swabs from chickens from four poultry farms was applied to the multiplex PCR assay. As a result, vaccine strains were detected in vaccinated farms, and wild-type strains were detected in unvaccinated farms (Figure 5).
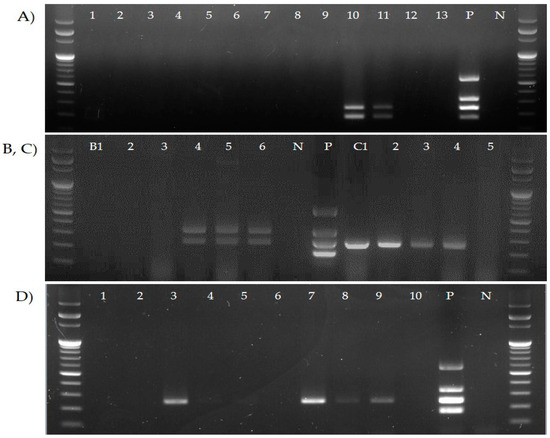
Figure 5.
Efficiency of the newly developed multiplex PCR assay on clinical samples. (A) Laying hen farm vaccinated with ts-11, (B) broiler breeder farm (no. B1–6) vaccinated with F strain, and (C,D) non-vaccinated broiler breeder farm (no. C1–5 and D1–10).
4. Discussion
MG is considered to be an important bacterial pathogen causing respiratory disease in chickens [4,15]. Once the flocks on farms are infected with this pathogen, they cannot be easily replaced with mycoplasma-free flocks. In particular, MG continues to appear on farms with poor sanitation or dirty environments and on farms of multi-age herds. The use of MG vaccines is perhaps the most effective and efficient means of preventing poultry farms from exposure to the aforementioned risk factors. In particular, the three kinds (ts-11, 6/85, and F strain) of MG vaccines have been widely used in many other countries, including the Netherlands, Asia (South Korea, Thailand, and Jordan), Australia, and Latin America (Mexico and Brazil) [3,14,15,16,17,18].
However, farm managers administer antibiotics to maintain vaccinated flocks or control other respiratory diseases. For this reason, frequent administration of antibiotics may increase resistance rates or interfere with the survival of vaccine strains.
Therefore, early detection of MG-contaminated sources in poultry farms can play an important role in effectively maintaining healthy flocks [18]. The assays applied for monitoring MG currently include bacterial isolation and serological and genetic assays. However, serological methods widely used for diagnosis lack the ability to differentiate between antibodies elicited by natural infection and those elicited by vaccination in vaccinated flocks. In comparison, culture for MG isolation can require at least 21 days and may be inhibited due to the rapid proliferation of other bacteria. Molecular genetic assays (GTS and MLST) could help effectively distinguish the three vaccine strains through sequencing, but doing so takes a long time [13,14,19]. Additionally, conventional PCR and real-time PCR are frequently used instead of culture to detect specific avian mycoplasma DNA, but these two PCR methods can only detect avian mycoplasma species or allow for the simultaneous detection of MG and MS [3,10,20]. Therefore, the new multiplex PCR assay in this study was developed to quickly and simply detect the three MG vaccine strains and wild-type strains simultaneously. Four target genes, the parE gene of MG species, the RS03710-rlmB interspace section of ts-11, the scpA gene of 6/85, and the MGF_RS03965 region of F strain, were specifically identified by the multiplex PCR assay in this study. The detection limit of our novel multiplex PCR assay was very low at 250 fg. Previous reports showed that the detection limit of real-time PCR described by Kahya et al. was 0.9 pg/μL [10]. However, the detection limit of a duplex PCR assay for MG and MS reported by Yadav et al. was 125 ng/mL [20]. Our novel multiplex PCR assay showed a lower detection limit than the real-time PCR and the duplex PCR methods reported previously. This multiplex PCR assay was optimized with a specialized PCR enzyme used in this study. The use of other PCR mixes diminishes sensitivity for a higher detection limit.
In conclusion, the newly developed multiplex PCR technique could be a very useful tool for finding field strains in vaccinated flocks. Moreover, this assay may be helpful as a tool to track specific immunity to Mycoplasma gallisepticum. In addition, this multiplex PCR method can replace the existing GTS technique because it can differentiate MG strains within a few hours using four primer sets.
Author Contributions
Data curation, S.-I.K.; writing—original draft preparation, S.-I.K.; validation, O.-M.L.; methodology, H.-J.L.; project administration, Y.-K.K.; resources, M.J.C.; investigation, J.-Y.J.; writing—review and editing, M.-S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant from the Animal and Plant Quarantine Agency (APQA) of South Korea [Grant number B-1543084–2020-22–01]; Animal and Plant Quarantine Agency: [Grant number B-1543084-2023-25-01].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Chung-Hyun Kim for providing the tracheas. The authors also thank Ok-Mi Jeong for providing the genomic DNA of the M. gallispeticum strains. All authors have read and consented to the final version of this manuscript.
Conflicts of Interest
The authors declare no potential conflict of interest.
References
- Swayne, D.E.; Boulianne, M.; Logue, C.M.; McDougald, L.R.; Nair, V.; Suarez, D.L. Diseases of Poultry, 14th ed.; Wiley Blackwell: Hoboken, NJ, USA, 2020; Volume 2, pp. 907–923. [Google Scholar]
- Emam, M.; Hashem, Y.M.; El-Hariri, M.; El-Jakee, J. Detection and antibiotic resistance of Mycoplasma gallisepticum and Mycoplasma synoviae among chicken flocks in Egypt. Vet. World 2020, 13, 1410–1416. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Available online: http://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access (accessed on 1 December 2022).
- Yadav, J.P.; Tomar, P.; Singh, Y.; Khurana, S.K. Insights on Mycoplasma gallisepticum and Mycoplasma synoviae infection in poultry: A systematic review. Anim. Biotechnol. 2021, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Noormohammadi, A.H.; Whithear, K.G. Comparision of the short-term and long-term efficacies of the Mycoplasma gallisepticum vaccines ts-11 and 6/85Comparision of the short-term and long-term efficacies of the Mycoplasma gallisepticum vaccines ts-11 and 6/85. Avian. Pathol. 2019, 48, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Ishfaq, M.; Hu, W.; Khan, M.Z.; Ahmad, I.; Guo, W.; Li, J. Current status of vaccine research, development, and challenges of vaccines for Mycoplasma gallisepticum. Poult. Sci. 2020, 99, 4195–4202. [Google Scholar] [CrossRef] [PubMed]
- Galluzzo, P.; Migliore, S.; Galuppo, L.; Condorelli, L.; Hussein, H.A.; Licitra, F.; Coltraro, M.; Sallemi, S.; Antoci, F.; Cascone, G.; et al. First molecular survey to detect Mycoplasma gallisepticum and Mycoplasma synoviae in poultry farms in a strategic production district of Sicily (South-Italy). Anim. 2022, 12, 962. [Google Scholar] [CrossRef] [PubMed]
- Raviv, Z.; Callison, S.A.; Ferguson-Noel, N.; Kleven, S.H. Strain differentiating real-time PCR for Mycoplasma gallisepticum live vaccine evaluation studies. Vet. Microbiol. 2008, 129, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Raviv, Z.; Kleven, S.H. The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas. Avian. Dis. 2009, 53, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Kahya, S.; Temelli, S.; Eyigor, A.; Carli, K.T. Real-time PCR culture and serology for the diagnosis of Mycoplasma gallisepticum in chicken breeder flocks. Vet. Microbiol. 2010, 144, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.Z.; Rahman, M.M.; Sultana, S. Seroprevalence of Mycoplasma gallisepticum antibody by ELISA and serum plate agglutination test of laying chicken. Vet. World 2015, 8, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M.; Heep, D.; Sun, S.; Ikuta, N.; Levisohn, S.; Kleven, S.H.; García, M. Use of molecular diversity of Mycoplasma gallisepticum by gene-targeted sequence (GTS) and random amplified polymorphic DNA (RAPD) analysis for epidemiological studies. Microbiology 2005, 151, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Matucci, A.; Stefani, E.; Gastaldelli, M.; Rossi, I.; De Grandi, G.; Gyuranecz, M.; Catania, S. Molecular differentiation of Mycoplasma gallisepticum outbreaks: A last decade study on Italian farms using GTS and MLST. Vaccines 2020, 8, 665. [Google Scholar] [CrossRef] [PubMed]
- Limsatanun, A.; Pakpinyo, S.; Limpavithayakul, K.; Prasertsee, T. Targeted sequencing analysis of Mycoplasma gallisepticum isolates in chicken layer and breeder flocks in Thailand. Sci. Rep. 2022, 12, 9900. [Google Scholar] [CrossRef] [PubMed]
- Feberwee, A.; de Wit, S.; Dijkman, R. Clinical expression, epidemiology, and monitoring of Mycoplasma gallisepticum and Mycoplasma synoviae: An update. Avian. Pathol. 2022, 51, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Fraga, A.P.; de Vargas, T.; Ikuta, N.; Fonseca, A.S.K.; Celmer, Á.J.; Marques, E.K.; Lunge, V.R. A multiplex real-time PCR for detection of Mycoplasma gallisepticum and Mycoplasma synoviae in clinical samples from Brazilian commercial poultry flocks. Braz. J. Microbiol. 2013, 30, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Petrone-García, V.M.; Tellez-Isaias, G.; Alba-Hurtado, F.; Vuong, C.N.; Lopez-Arellano, R. Isolation and antimicrobial sensitivity of Mycoplasma synoviae and Mycoplasma gallisepticum from vaccinated hens in Mexico. Pathogens 2020, 9, 924. [Google Scholar] [CrossRef] [PubMed]
- Veen, C.T.; Dijkman, R.; de Wit, J.J.; Gyuranecz, M.; Feberwee, A. Decrease of Mycoplasma gallisepticum seroprevalence and introduction of new genotypes in Dutch commercial poultry during the years 2001–2018. Avian. Pathol. 2021, 50, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Ball, C.; Forrester, A.; Ganapathy, K. Co-circulation of genetically diverse population of vaccine related and unrelated respiratory mycoplasmas and viruses in UK poultry flocks with health or production problems. Vet. Microbiol. 2018, 225, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Yadav, J.P.; Singh, Y.; Jindl, N.; Mahajan, N.K. Rapid and specific detection of Mycoplasma gallisepticum and Mycoplasma synoviae infection in poultry using single and duplex PCR assays. J. Microbiol. Methods 2022, 192, 106365. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).