Gut Microbiota and COVID-19: Potential Implications for Disease Severity
Abstract
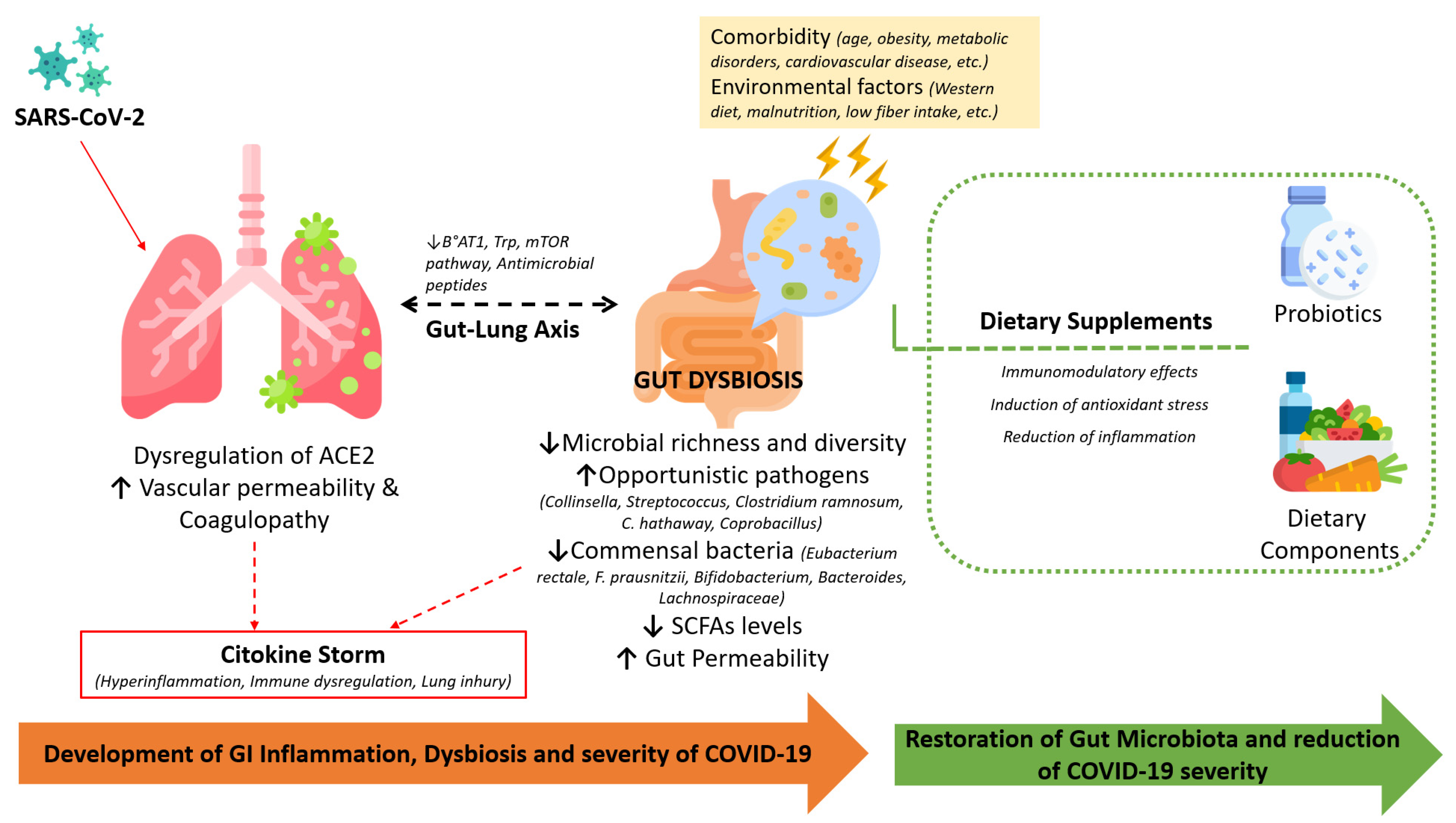
:1. Introduction
2. SARS-CoV-2 Pandemic
Inflammation and COVID-19
3. The Gastrointestinal Tract and COVID-19 Infection
3.1. Gastrointestinal Clinical Manifestations and Abnormalities
3.2. The Gut Microbiome and Its Role in COVID-19 Disease
| Population and Characteristics | Methods | Main Results | Ref. |
|---|---|---|---|
| 62 COVID-19 patients | Next-generation sequencing of the V4 region of the 16S ribosomal RNA gene. Samples from HC, severe COVID-19, and seasonal Flu patients were collected at the first visit to the hospital. | Infected patients showed: in alpha diversity abundance of Streptococcus, Clostridium, Lactobacillus, and Bifidobacterium Bacteroides, Roseburia, Faecalibacterium, Coprococcus, and Parabacteroides sera levels of IL-18 | [48] |
| 33 seasonal flu patients (Flu) 40 healthy controls (HC) | |||
| 30 COVID-19 positive patients. (10 patients with comorbidities) | 16S rRNA gene sequencing. Stool samples were collected upon admission. | Compared to controls, COVID-19 patients had lymphocyte counts and levels of IL-6 and TNF-α bacterial diversity, abundance of Ruminococcaceae and Lachnospiraceae familyies abundance of opportunistic pathogens: Streptococcus, Rothia, Veillonella, and Actinomyces. A positive correlation was described between PCR levels and opportunistic bacteria. | [49] |
| 2 groups: 30 healthy controls 24 H1N1 flu patients | |||
| 15 COVID-19 positive patients | Whole-genome sequencing. Stool samples were collected 2-3 times per week during hospitalization. | An in Collinsella aerofaciens was detected in fecal samples, with high infectivity. In contrast, fecal samples with low SARS-CoV-2 infectivity had levels of Parabacteroides, Bacteroides and Lachnospiraeae. Coprobacillus, Clostridium ramosum, and C. hathewayi in the Firmicutes phylum were the best bacteria, showing a positive correlation with COVID-19 severity. | [64] |
| 15 healthy controls | |||
| 15 COVID-19 positive patients | Whole-genome sequencing. Samples were collected 2-3 times per week during hospitalization. | COVID-19 patients on admission, compared to healthy controls, had numbers of Clostridium hathewayi, Actinomyces viscosus, and Bacteroides nordii. Alistipes onderdonkii and Faecalibacterium prausnitzii were negatively correlated with severe COVID-19. | [44] |
| 15 healthy controls | |||
| 117 patients infected with SARS-CoV-2 | 16S rRNA gene sequencing of the V3-V4 region. | In total, virus-positive patients showed a: bacterial richness; Actinobacteria, Bifidobacterium, Streptococcus, and Collinsella genera; Proteobacteria, Bacteroidetes, Enterobacteriaceae, and Bacteroides genera. Patients with severe COVID-19 exhibited: blood levels of inflammatory markers; CD8+ T cell number; abundance of F. prausnitzii and Roseburia. | [51] |
| 95 SARS-CoV-2 negative patients | |||
| 50 COVID-19 positive patients, symptomatic (mild, moderate, severe) and asymptomatic | Shotgun next-generation sequencing was performed. Samples were collected within 48 h after the onset of symptoms or within one week after positivity. | Gut microbiota of 28 severely symptomatic SARS-CoV-2 patients had bacterial α-diversity, levels of the protective Bifidobacterium, Faecalibacterium, and Roseburium genera, and abundance of Bacteroides. | [54] |
| 20 COVID-19 negative controls; exposed controls |
3.3. Probiotic Supplementations: Possible Strategy for Modulating the Gut Microbiota in COVID-19
4. Nutrition and COVID-19: An Evidence-Based Overview of Risk Factors and Protective Factors
4.1. Implications of Metabolic Alterations and Malnutrition
4.2. Role of Nutrients and Bioactive Components
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef]
- Cyprian, F.; Sohail, M.U.; Abdelhafez, I.; Salman, S.; Attique, Z.; Kamareddine, L.; Al-Asmakh, M. SARS-CoV-2 and immune-microbiome interactions: Lessons from respiratory viral infections. Int. J. Infect. Dis. 2021, 105, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Lantinga, M.A.; Mönkemüller, K. COVID-19 in gastroenterology and hepatology: Where will we be? United Eur. Gastroenterol. J. 2021, 9, 743–744. [Google Scholar] [CrossRef]
- Meringer, H.; Mehandru, S. Gastrointestinal post-acute COVID-19 syndrome. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 345–346. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in the microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef]
- Yang, X.L. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2016, 90, 3253–3256. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Xavier, J.; Giovanetti, M.; Adelino, T.; Fonseca, V.; Barbosa da Costa, A.V.; Ribeiro, A.A.; Felicio, K.N.; Duarte, C.G.; Ferreira Silva, M.V.; Salgado, Á.; et al. The ongoing COVID-19 epidemic in Minas Gerais, Brazil: Insights from epidemiological data and SARS-CoV-2 whole genome sequencing. Emerg. Microbes Infect. 2020, 9, 1824–1834. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, s.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Cella, E.; Benedetti, F.; Rife, M.B.; Fonseca, V.; Fabris, S.; Campisi, G.; Ciccozzi, A.; Angeletti, S.; Borsetti, A.; et al. SARS-CoV-2 shifting transmission dynamics and hidden reservoirs potentially limit the efficacy of public health interventions in Italy. Commun. Biol. 2021, 4, 489. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- CDC, COVID-19 Data. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed on 1 July 2022).
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154, October, 2020. Erratum in Nat. Rev. Microbiol. 2022, 20, 315. [Google Scholar] [CrossRef] [PubMed]
- Hindson, J. COVID-19: Faecal-oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef]
- Kaźmierczak-Siedlecka, K.; Vitale, E.; Makarewicz, W. COVID-19-gastrointestinal and gut microbiota-related aspects. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10853–10859. [Google Scholar]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Giovanetti, M.; Slavov, S.N.; Fonseca, V.; Wilkinson, E.; TegallyLegally, H.; Patané, J.S.L.; Viala, V.L.; San, J.E.; Rodrigues, E.S.; Santos, E.V.; et al. Genomic epidemiology reveals the impact of national and international restrictions measures on the SARS-CoV-2 epidemic in Brazil. medRxiv, 2022; Preprint. [Google Scholar]
- WHO. Variants Tracking. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 1 July 2022).
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses Methods Protoc. 2015, 1282, 1–23. [Google Scholar]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, M.; Chen, X.; Montaner, L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc Biol. 2020, 108, 17–41. [Google Scholar] [CrossRef] [PubMed]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; van de Veen, W.; Brüggen, M.-C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef]
- Teuwen, L.A.; Geldhof, V.; Pasut, A.; Carmeliet, P. COVID-19: The vasculature unleashed. Nat. Rev. Immunol. 2020, 20, 389. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.P.; Patel, P.A.; Vunnam, R.R.; Hewlett, A.T.; Jain, R.; Jing, R.; Vunnam, S.R. Gastrointestinal, hepatobiliary, and pancreatic manifestations of COVID-19. J. Clin. Virol. 2020, 128, 104386. [Google Scholar] [CrossRef]
- Mehandru, S.; Merad, M. Pathological sequelae of long-haul COVID. Nat. Immunol. 2022, 23, 194–202. [Google Scholar] [CrossRef]
- Leung, W.K.; To, K.F.; Chan, P.K. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 2003, 125, 1011–1017. [Google Scholar] [CrossRef]
- Tariq, R.; Saha, S.; Furqan, F.; Hassett, L.; Pardi, D.; Khanna, S. Prevalence and Mortality of COVID-19 Patients with Gastrointestinal Symptoms: A Systematic Review and Meta-analysis. Mayo Clin. Proc. 2020, 95, 1632–1648. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.; Chan, P.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Blackett, J.W.; Wainberg, M.; Elkind, M.S.V.; Freedberg, D.E. Potential long coronavirus disease 2019 gastrointestinal symptoms 6 months after coronavirus infection are associated with mental health symptoms. Gastroenterology 2022, 162, 648–650.e2. [Google Scholar] [CrossRef]
- Azouz, E.; Yang, S.; Monnier-Cholley, L.; Arrivé, L. Systemic arterial thrombosis and acute mesenteric ischemia in a patient with COVID-19. Intensive Care Med. 2020, 46, 1464–1465. [Google Scholar] [CrossRef]
- Chan, K.H.; Poon, L.L.; Cheng, V.C. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 2004, 10, 294–299. [Google Scholar] [CrossRef]
- Wurtz, N.; Penant, G.; Jardot, P.; Duclos, N.; La Scola, B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 477–484. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.; Wang, Y.; Yuen, T.T.; Chai, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; Zhang, X.; et al. SARS-CoV-2 Induces a More Robust Innate Immune Response and Replicates Less Efficiently Than SARS-CoV in the Human Intestines: An Ex Vivo Study with Implications on Pathogenesis of COVID-19. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 771–781. [Google Scholar] [CrossRef]
- De Maio, F.; Lo Cascio, E.; Babini, G.; Sali, M.; Della Longa, S.; Tilocca, B.; Roncada, P.; Arcovito, A.; Sanguinetti, M.; Scambia, G.; et al. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020, 22, 592–597. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, C.; Zhang, S.; Duan, C.; Shang, H.; Bai, T.; Hou, X. Diarrhea and altered inflammatory cytokine pattern in severe coronavirus disease 2019: Impact on disease course and in-hospital mortality. J. Gastroenterol. Hepatol. 2021, 36, 421–429. [Google Scholar] [PubMed]
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 Imbalance as a Key Player for the Poor Outcomes in COVID-19 Patients with Age-Related Comorbidities-Role of Gut Microbiota Dysbiosis. Ageing Res. Rev. 2020, 62, 101123. [Google Scholar] [CrossRef]
- De Oliveira, A.P.; Lopes, A.L.F.; Pacheco, G.; de Sa Guimaraes Noleto, I.R.; Nicolau, L.A.D.; Medeiros, J.V.R. Premises among SARS-CoV-2, dysbiosis and diarrhea: Walking through the ACE2/mTOR/autophagy route. Med. Hypotheses. 2020, 144, 110243. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 25, 477–481, 487. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012, 3, 463. [Google Scholar] [CrossRef]
- Fagundes, C.T. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in Germfree Mice. J. Immunol. 2012, 188, 1411–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panebianco, C.; Eddine, F.B.N.; Forlani, G.; Palmieri, G.; Tatangelo, L.; Villani, A.; Xu, L.; Accolla, R.; Pazienza, V. Probiotic Bifidobacterium lactis, anti-oxidant vitamin E/C and anti-inflammatory dha attenuate lung inflammation due to pm2.5 exposure in mice. Benef. Microbes 2019, 10, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Mageswary, M.U.; Ang, X.Y.; Lee, B.K.; Chung, Y.-L.F.; Azhar, S.N.A.; Hamid, I.J.A.; Bakar, H.A.; Roslan, N.S.; Liu, X.; Kang, X.; et al. Probiotic Bifidobacterium lactis Probio-M8 treated and prevented acute RTI, reduced antibiotic use and hospital stay in hospitalized young children: A randomized, double-blind, placebo-controlled study. Eur. J. Nutr. 2022, 61, 1679–1691. [Google Scholar] [CrossRef]
- Kageyama, Y.; Nishizaki, Y.; Aida, K.; Yayama, K.; Ebisui, T.; Akiyama, T.; Nakamura, T. Lactobacillus plantarum induces innate cytokine responses that potentially provide a protective benefit against COVID-19: A single-arm, double-blind, prospective trial combined with an in vitro cytokine response assay. Exp. Ther. Med. 2022, 23, 18–20. [Google Scholar] [CrossRef]
- Tao, W.; Zhang, G.; Wang, X.; Guo, M.; Zeng, W.; Xu, Z.; Gao, D.; Pan, A.; Wang, Y.; Zhang, K.; et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microbiol. 2020, 5, 100023. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin. Infect. Dis. 2020, 159, 944–995. [Google Scholar]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K.; et al. Depicting SARS-CoV-2 fecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2020, 159, 944–995. [Google Scholar]
- Reinold, J.; Farahpour, F.; Fehring, C.; Wolff, S.; Konik, M.; Korth, J.; van Baal, L.; Hoffmann, D.; Buer, J.; Witzke, O.; et al. A Pro-Inflammatory Gut Microbiome Characterizes SARS-CoV-2 Infected Patients and a Reduction in the Connectivity of an Anti-Inflammatory Bacterial Network Associates with Severe COVID-19. Front. Cell Infect. Microbiol. 2021, 11, 747816. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L. Mining the human gut microbiota for immunomodulatory organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.Y.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients with COVID-19. Gastroenterology 2022, 162, 548–561.e4. [Google Scholar] [CrossRef]
- Yu, L.; Tong, Y.; Shen, G.; Fu, A.; Lai, Y.; Zhou, X.; Yuan, Y.; Wang, Y.; Pan, Y.; Yu, Z.; et al. Immunodepletion with hypoxemia: A potentially high-risk subtype of coronavirus disease 2019. medRxiv 2020, 16-99-142. [Google Scholar] [CrossRef]
- Ganesan, K.; Chung, S.K.; Vanamala, J.; Xu, B. Causal relationship between diet-induced gut microbiota changes and diabetes: A novel strategy to transplant Faecalibacterium prausnitzii in preventing diabetes. Int. J. Mol. Sci. 2018, 159, 3720. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.W.; Roberts, A.P.; Frugé, A.D. Negative association between Mediterranean diet adherence and COVID-19 cases and related deaths in Spain and 23 OECD countries: An ecological study. Front. Nutr. 2021, 8, 591964. [Google Scholar] [CrossRef]
- Nishino, K.; Imaeda, H.; Sakai, S.; Ohno, M.; Nishida, A.; Andoh, A. The abundance of Clostridium hathewayi, a potent inducer of t helper 17 (Th17) cells, is associated with the disease severity of Crohn’s disease. Gastroenterology 2017, 152, S993. [Google Scholar] [CrossRef]
- Shinagawa, N.; Taniguchi, M.; Hirata, K.; Furuhata, T.; Fukuhara, K.; Mizugucwi, T.; Osanai, H.; Yanai, Y.; Hata, F.; Kihara, C.; et al. Bacteria isolated from surgical infections and its susceptibilities to antimicrobial agents--special references to bacteria isolated between April 2010 and March 2011. Jpn. J. Antibiot. 2014, 67, 293–334. [Google Scholar]
- Gottschalk, G.; Knox, K.; Roy, A. ACE2: At the Crossroad of COVID-19 and Lung Cancer. Gene Rep. 2021, 23, 101077. [Google Scholar] [CrossRef]
- Sodhi, C.P.; Nguyen, J.; Yamaguchi, Y.; Werts, A.D.; Lu, P.; Ladd, M.R.; Fulton, W.B.; Kovler, M.L.; Wang, S.; Prindle, T.; et al. A Dynamic Variation of Pulmonary ACE2 Is Required to Modulate Neutrophilic Inflammation in Response to Pseudomonas Aeruginosa Lung Infection in Mice. J. Immunol. 2019, 203, 3000–3012. [Google Scholar] [CrossRef] [PubMed]
- De, R.; Dutta, S. Role of the Microbiome in the Pathogenesis of COVID-19. Front. Cell Infect. Microbiol. 2022, 12, 736397. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Michelle, G.R.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.-L.; Salomé-Desnoulez, S.; Deryuter, L.; et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020, 30, 2934–2947. [Google Scholar] [CrossRef]
- Sokol, H.; Contreras, V.; Maisonnasse, P.; Desmons, A.; Delache, B.; Sencio, V.; Machelart, A.; Brisebarre, A.; Humbert, L.; Deryuter, L.; et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes. 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Sencio, V.; Gallerand, A.; Gomes Machado, M.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c- Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 2018, 48, 992–1005.e8. [Google Scholar] [CrossRef]
- Brown, J.A.; Sanidad, K.Z.; Lucotti, S.; Lieber, C.M.; Cox, R.M.; Ananthanarayanan, A.; Basu, S.; Chen, J.; Shan, M.; Amir, M.; et al. Gut microbiota-derived metabolites confer protection against SARS-CoV-2 infection. Gut Microbes. 2022, 14, 2105609. [Google Scholar] [CrossRef]
- Giovanetti, M.; Slavov, S.N.; Fonseca, V.; Wilkinson, E.; Tegally, H.; Patané, J.S.L.; Viala, V.L.; San, E.J.; Rodrigues, E.S.; Santos, E.V.; et al. Genomic epidemiology of the SARS-CoV-2 epidemic in Brazil. Nat. Microbiol. 2022, 7, 1490–1500. [Google Scholar] [CrossRef]
- Lau, A.S.; Yanagisawa, N.; Hor, Y.Y.; Lew, L.C.; Ong, J.S.; Chuah, L.O.; Lee, Y.Y.; Choi, S.B.; Rashid, F.; Wahid, N.; et al. Bifidobacterium longum BB536 alleviated upper respiratory illnesses and modulated gut microbiota profiles in Malaysian pre-school children. Benef Microbes. 2018, 9, 61–70. [Google Scholar] [CrossRef]
- Chong, H.X.; Yusoff, N.A.A.; Hor, Y.Y.; Lew, L.C.; Jaafar, M.H.; Choi, S.B.; Yusoff, M.S.B.; Wahid, N.; Abdullah, M.F.I.L.; Zakaria, N.; et al. Lactobacillus plantarum DR7 improved upper respiratory tract infections via enhancing immune and inflammatory parameters: A randomized, double-blind, placebo-controlled study. J. Dairy Sci. 2019, 102, 4783–4797. [Google Scholar] [CrossRef] [PubMed]
- Yan, -Y.H.; Lee, -C.L.; Amy, S.-Y.L.; Jia, -S.O.; Li, -O.C.; Yeong, -Y.L.; Sy, -B.C.; Faridah, R.; Normala, W.; Sun, Z.; et al. Probiotic Lactobacillus casei Zhang (LCZ) alleviates respiratory, gastrointestinal & RBC abnormality via immune-modulatory, anti-inflammatory & anti-oxidative action. J. Funct. Foods 2018, 44, 235–245. [Google Scholar]
- Ivashkin, V.; Fomin, V.; Moiseev, S.; Brovko, M.; Maslennikov, R.; Ulyanin, A.; Sholomova, V.; Vasilyeva, M.; Trush, E.; Shifrin, O.; et al. Efficacy of a Probiotic Consisting of Lacticaseibacillus rhamnosus PDV 1705, Bifidobacterium bifidum PDV 0903, Bifidobacterium long subs. infantis PDV 1911, and Bifidobacterium longum subsp. lolongDV 2301 in the Treatment of Hospitalized Patients with COVID-19: A Randomized Controlled Trial. Probiotics Antimicrob. Proteins. 2021, 36, 1–9. [Google Scholar]
- Gutiérrez-Castrellón, P.; Gandara-Martí, T.; Abreu, A.T.A.Y.; Nieto-Rufino, C.D.; López-Orduña, E.; Jiménez-Escobar, I.; Jiménez-Gutiérrez, C.; López-Velazquez, G.; Espadaler-Mazo, J. Probiotic improves symptomatic and viral clearance in COVID-19 outpatients: A randomized, quadruple-blinded, placebo-controlled trial. Gut Microbes. 2022, 14, 2018899. [Google Scholar] [CrossRef]
- People at Higher Risk from Coronavirus (COVID-19). Available online: https://www.nhs.uk/conditions/coronavirus-covid-19/people-at-higher-risk/ (accessed on 18 July 2022).
- Kassi, E.; Pervanidou, P.; Kaltsas, G.; Chrousos, G. Metabolic Syndrome: Definitions and Controversies. BMC Med. 2011, 9, 48. [Google Scholar] [CrossRef]
- Davenport, C.; Finucane, F.M. Coronavirus and Obesity: Could Insulin Resistance Mediate the Severity of COVID-19 Infection? Front. Public Health 2020, 8, 184. [Google Scholar]
- COVID-ICU Group on behalf of the REVA Network and the COVID-ICU Investigators. Clinical characteristics and day-90 outcomes of 4244 critically ill adults with COVID-19: A prospective cohort study. Intensive Care Med. 2021, 47, 60–73. [Google Scholar] [CrossRef]
- Dalan, R.; Bornstein, S.R.; El-Armouche, A.; Rodionov, R.N.; Markov, A.; Wielockx, B.; Beuschlein, F.; Boehm, B.O. The ACE-2 in COVID-19: Foe or Friend? HormMetab. Res. 2020, 52, 257–263. [Google Scholar] [CrossRef]
- Jia. Hongpeng Pulmonary Angiotensin-Converting Enzyme 2 (ACE2) and Inflammatory Lung Disease. SHOCK 2016, 46, 239–248. [CrossRef]
- Dietz, W.; Santos-Burgoa, C. Obesity and its implications for COVID-19 mortality. Obesity 2020, 28, 1005. [Google Scholar] [CrossRef] [PubMed]
- Geurts, L.; Neyrinck, A.M.; Delzenne, N.M.; Knauf, C.; Cani, P.D. Gut Microbiota Controls Adipose Tissue Expansion, Gut Barrier and Glucose Metabolism: Novel Insights Into Molecular Targets and Interventions Using Prebiotics. Benef. Microbes 2014, 5, 3–17. [Google Scholar] [CrossRef]
- Sencio, V.; Benech, N.; Robil, C.; Deruyter, L.; Heumel, S.; Machelart, A.; Sulpice, T.; Lamazière, A.; Grangette, C.; Briand, F.; et al. Alteration of the gut microbiota’s composition and metabolic output correlates with COVID-19-like severity in obese NASH hamsters. Gut Microbes 2022, 14, 2100200. [Google Scholar] [CrossRef] [PubMed]
- Suratt, P.M.; Wilhoit, S.C.; Hsiao, H.S.; Atkinson, R.L.; Rochester, D.F. Compliance of Chest Wall in Obese Subjects. J. Appl. Physiol. 1984, 57, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Inuma, K. Low COVID-19 Infection and Mortality in Rice Eating Countries. Sch. J. Food Nutr. 2020, 3, 326–328. [Google Scholar] [CrossRef]
- Laviano, A.; Koverech, A.; Zanetti, M. Nutrition Support in the Time of SARS-CoV-2 (COVID-19). Nutrition 2020, 74, 110834. [Google Scholar] [CrossRef]
- Yaqoob, P. Ageing Alters the Impact of Nutrition on Immune Function. Proc. Nutr. Soc. 2017, 76, 347–351. [Google Scholar] [CrossRef]
- Singer, P.; Blaser, A.R.; Berger, M.M.; Alhazzani, W.; Calder, P.C.; Casaer, M.P.; Hiesmayr, M.; Mayer, K.; Montejo, J.C.; Pichard, C.; et al. ESPEN Guideline on Clinical Nutrition in the Intensive Care Unit. Clin. Nutr. 2019, 38, 48–79. [Google Scholar] [CrossRef]
- Trompette, A. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y. Potential interventions for novel coronavirus in China: A systematic review. J. Med. Virol. 2020, 92, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, A.; Infante, M.; Ricordi, C. Editorial—Vitamin D Status: A Key Modulator of Innate Immunity and Natural Defense from Acute Viral Respiratory Infections. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4048–4052. [Google Scholar] [PubMed]
- Sadeghi, K.; Wessner, B.; Laggner, U.; Ploder, M.; Tamandl, D.; Friedl, J. Vitamin D3 down-regulates monocyte TLR expression and triggers hyporesponsiveness to pathogen-associated molecular patterns. Eur. J. Immunol. 2006, 36, 361–370. [Google Scholar] [CrossRef]
- Belancic, A.; Kresovic, A.; Racki, V. Potential pathophysiological mechanisms leading to increased COVID-19 susceptibility and severity in obesity. Obes Med. 2020, 19, 10–259. [Google Scholar]
- Grant, W.B.; Lahore, H.; McDonnell, S.L.; Baggerly, C.A.; French, C.B.; Aliano, J.L.; Bhattoa, H.P. Evidence That Vitamin D Supplementation Could Reduce Risk of Influenza and COVID-19 Infections and Deaths. Nutrients 2020, 12, 988. [Google Scholar] [CrossRef]
- Skrajnowska, D.; Brumer, M.; Kankowska, S.; Matysek, M.; Miazio, N.; Bobrowska-Korczak, B. COVID-19: Diet Composition and Health. Nutrients 2021, 13, 2980. [Google Scholar] [CrossRef]
- Carr, A.C.; Maggini, S. Vitamin, C and immune function. Nutrients 2017, 9, 1211. [Google Scholar] [CrossRef]
- Choe, J.-Y.; Kim, S.-K. Quercetin and Ascorbic Acid Suppress Fructose-Induced NLRP3 Inflammasome Activation by Blocking Intracellular Shuttling of TXNIP in Human Macrophage Cell Lines. Inflammation 2017, 40, 980–994. [Google Scholar] [CrossRef]
- Di Renzo, L.; Gualtieri, P.; Pivari, F.; Soldati, L.; Attinà, A.; Leggeri, C.; Cinelli, G.; Tarsitano, M.G.; Caparello, G.; Carrano, E.; et al. COVID-19: Is there a role for immunonutrition in the obese patient? J. Transl Med. 2020, 18, 415–800. [Google Scholar] [CrossRef]
- Allen, L.; Benoist, B.D.; Dary, O.; Hurrell, R. WHO and FAO of the United Nations. In Guidelines on Food Fortification with Micronutrients; World Health Organization: Geneva, Switzerland; Food and Agriculture Organization: Rome, Italy; United Nations: San Francisco, CA, USA, 2006. [Google Scholar]
- Huang, Z.; Liu, Y.; Qi, G.; Brand, D.; Zheng, S.G. Role of vitamin A in the immune system. J. Clin. Med. 2018, 7, 258. [Google Scholar] [CrossRef]
- Morais, A.H.d.A.; Passos, T.S.; Vale, S.H.d.L.; Maia, J.K.d.S.; Maciel, B.L.L. Obesity and the increased risk for COVID-19: Mechanisms and nutritional management. Nutr. Res. Rev. Vol. 2021, 32, 209–221. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Van den Worm, S.H.; Sims, A.C.; Baric, R.S.; Snijder, E.J.; Van Hemert, M.J. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010, 6, e1001176. [Google Scholar] [CrossRef]
- Saeed, F.; Nadeem, M.; Ahmed, R.S.; Tahir Nadeem, M.; Arshad, M.S.; Ullah, A. Studying the impact of nutritional immunology underlying the modulation of immune responses by nutritional compounds—a review. Food Agric. Immunol. 2016, 27, 205–229. [Google Scholar] [CrossRef]
- Guarino, M.P.L.; Altomare, A.; Emerenziani, S.; Di Rosa, C.; Ribolsi, M.; Balestrieri, P.; Iovino, P.; Rocchi, G.; Cicala, M. Mechanisms of Action of Prebiotics and Their Effects on Gastro-Intestinal Disorders in Adults. Nutrients 2020, 12, 1037. [Google Scholar] [CrossRef]
- Thorburn, A.N.; Macia, L.; Mackay, C.R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014, 40, 833–842. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Das, U.N. Can Bioactive Lipids Inactivate Coronavirus (COVID-19)? Arch. Med. Res. 2020, 51, 282–286. [Google Scholar] [CrossRef]
- Simopoulos, A. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef]
- Radzikowska, U.; Rinaldi, A.O.; Çelebi Sözener, Z.; Karaguzel, D.; Wojcik, M.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The Influence of Dietary Fatty Acids on Immune Responses. Nutrients 2019, 11, 2990. [Google Scholar] [CrossRef]
- Calder, P.C. Dietary modification of inflammation with lipids. Proc. Nutr. Soc. 2002, 61, 345–358. [Google Scholar] [CrossRef]
- Asher, A.; Tintle, N.L.; Myers, M.; Lockshon, L.; Bacareza, H.; Harris, W.S. Blood Omega-3 Fatty Acids and Death from COVID-19: A Pilot Study. Prostaglandins Leukot. Essent. Fat. Acids 2021, 166, 102250. [Google Scholar] [CrossRef]
- Chiang, E.I.; Syu, J.N.; Hung, H.C.; Rodriguez, R.L.; Wang, W.J.; Chiang, E.R.; Chiu, S.C.; Chao, C.Y.; Tang, F.Y. N-3 polyunsaturated fatty acids block the trimethylamine-N-oxide-ACE2-TMPRSS2 cascade to inhibit the infection of human endothelial progenitor cells by SARS-CoV-2. J. Nutr. Biochem. 2022, 109, 109102. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchi, G.; Giovanetti, M.; Benedetti, F.; Borsetti, A.; Ceccarelli, G.; Zella, D.; Altomare, A.; Ciccozzi, M.; Guarino, M.P.L. Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens 2022, 11, 1050. https://doi.org/10.3390/pathogens11091050
Rocchi G, Giovanetti M, Benedetti F, Borsetti A, Ceccarelli G, Zella D, Altomare A, Ciccozzi M, Guarino MPL. Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens. 2022; 11(9):1050. https://doi.org/10.3390/pathogens11091050
Chicago/Turabian StyleRocchi, Giulia, Marta Giovanetti, Francesca Benedetti, Alessandra Borsetti, Giancarlo Ceccarelli, Davide Zella, Annamaria Altomare, Massimo Ciccozzi, and Michele Pier Luca Guarino. 2022. "Gut Microbiota and COVID-19: Potential Implications for Disease Severity" Pathogens 11, no. 9: 1050. https://doi.org/10.3390/pathogens11091050
APA StyleRocchi, G., Giovanetti, M., Benedetti, F., Borsetti, A., Ceccarelli, G., Zella, D., Altomare, A., Ciccozzi, M., & Guarino, M. P. L. (2022). Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens, 11(9), 1050. https://doi.org/10.3390/pathogens11091050











