Poxvirus Recombination
Abstract
1. Biological Function of Recombination
2. Poxviruses
3. Historical Insights
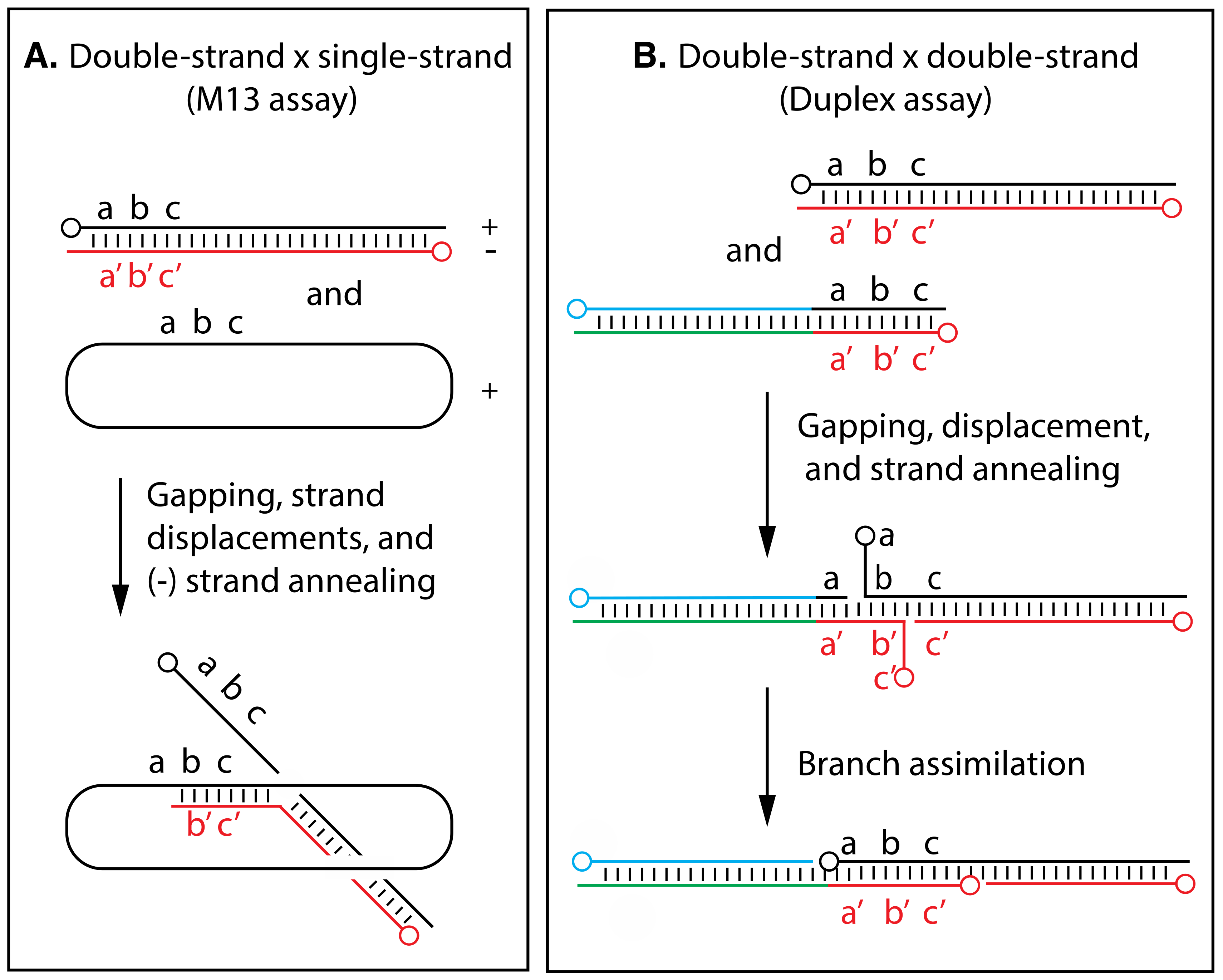
4. Marker Rescue Studies
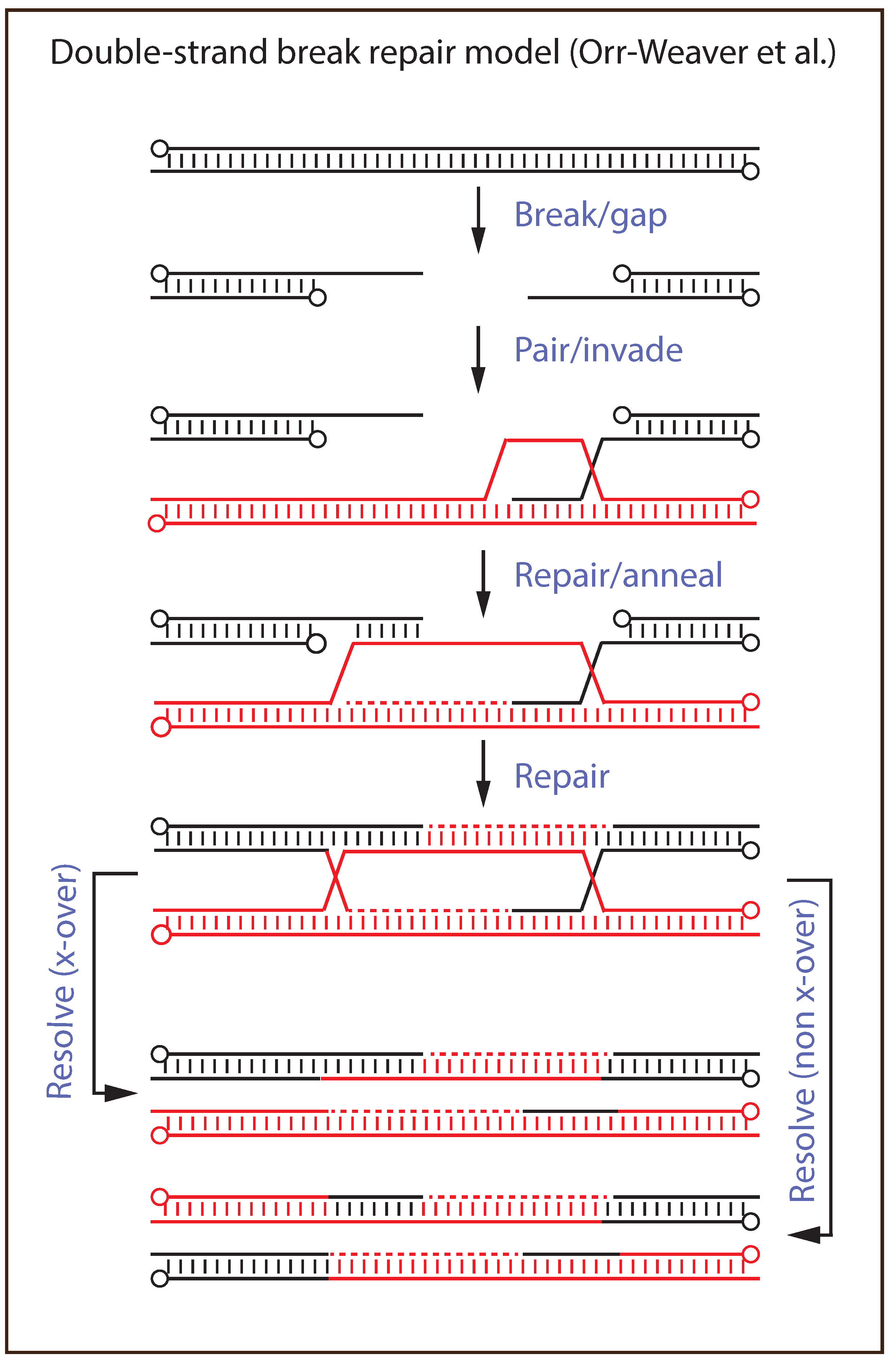
5. Recombination and Repair Models
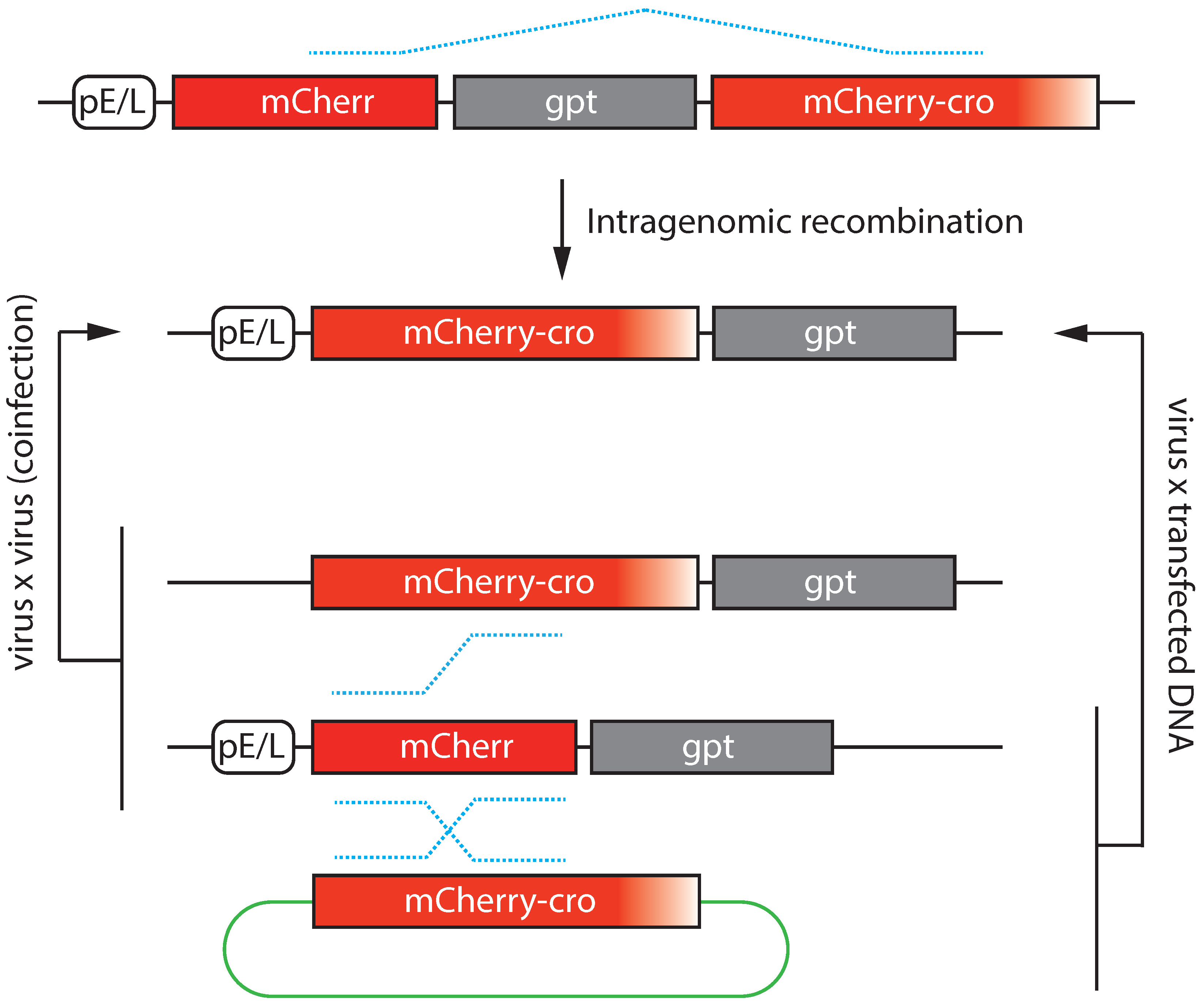
6. Transfection Studies
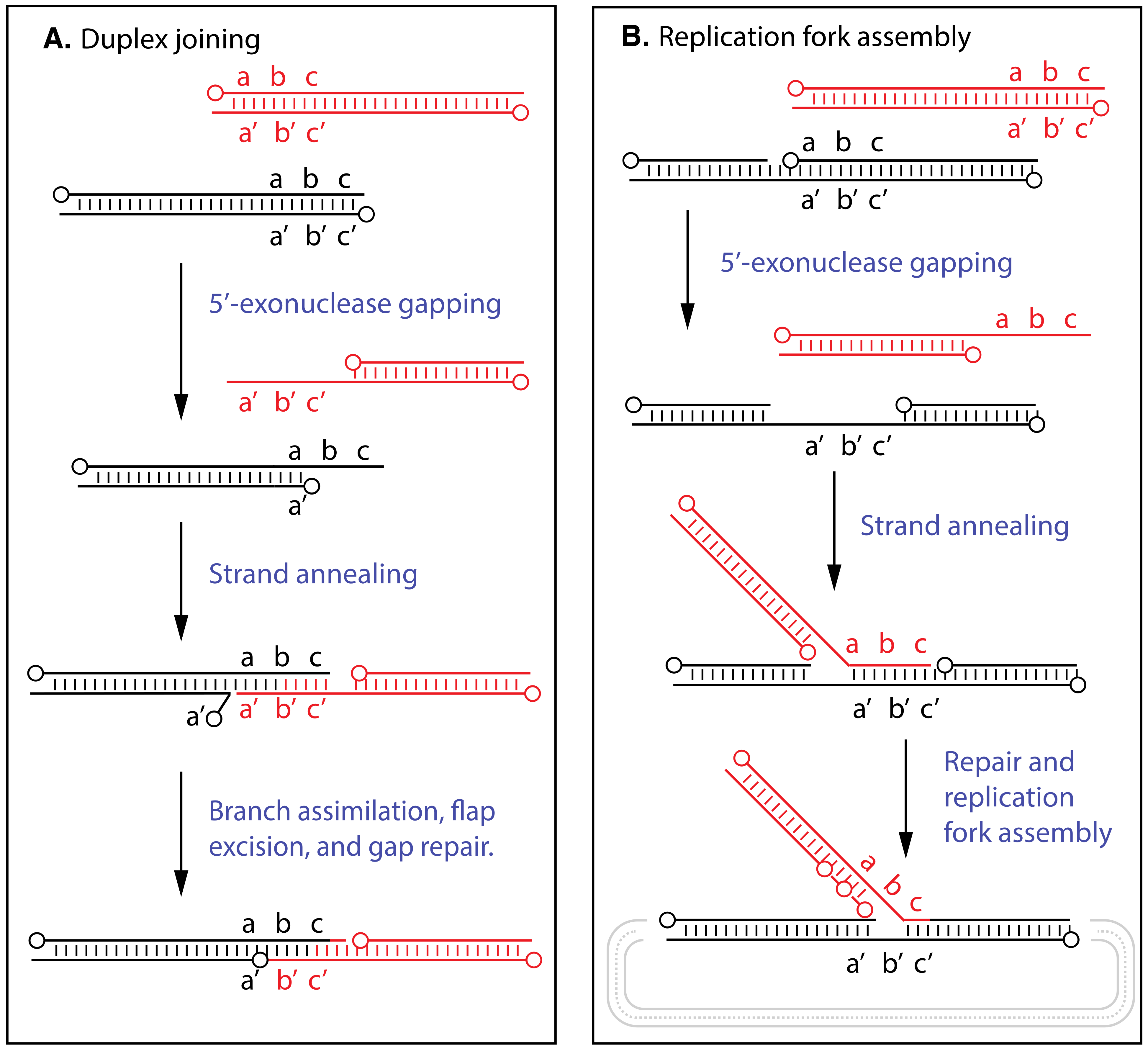
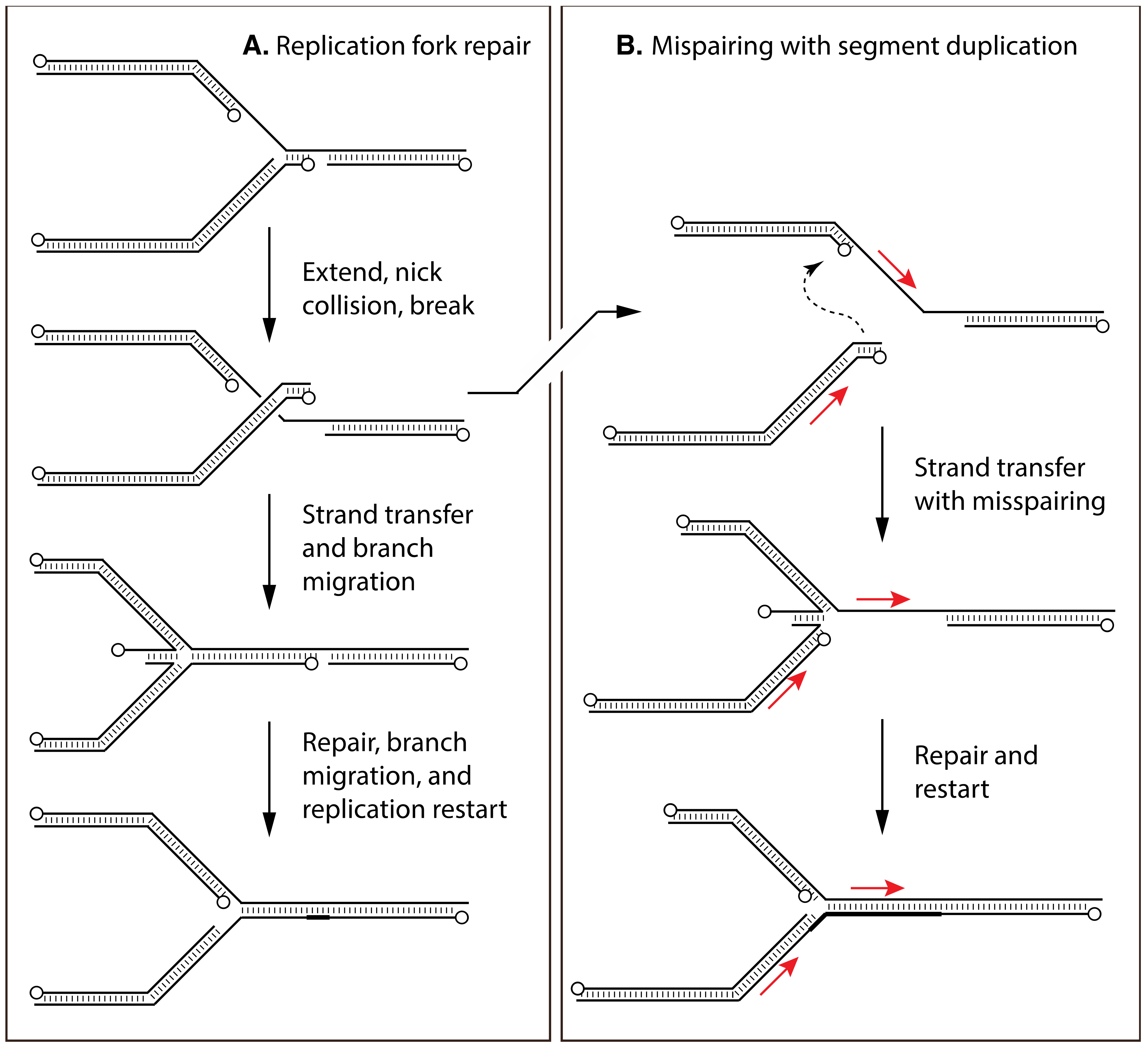
7. Biochemical Studies
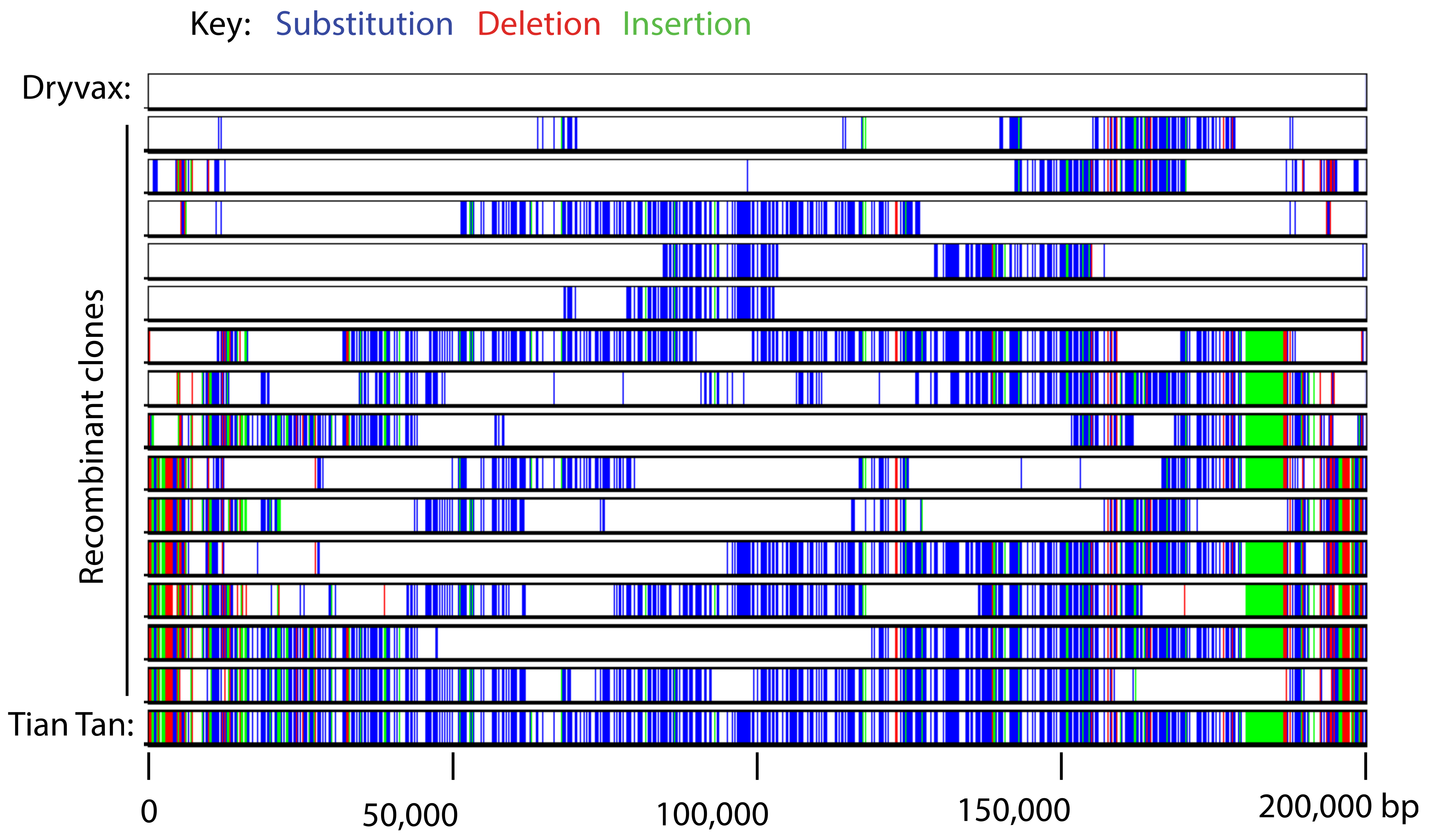
8. Sequencing and Genomics
9. How Does the Cellular Environment Influence These Events?
10. Applications and Implications
11. Unanswered Questions and Way(s) Forward
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koonin, E.V. The Origin at 150: Is a new evolutionary synthesis in sight? Trends Genet. 2009, 25, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. The evolutionary advantage of recombination. Genetics 1974, 78, 737–756. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.J.; Margulies, A.D. Isolation and Characterization of Recombination-Deficient Mutants of Escherichia Coli K12. Proc. Natl. Acad. Sci. USA 1965, 53, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Goodman, M.F.; Kreuzer, K.N.; Sherratt, D.J.; Sandler, S.J.; Marians, K.J. The importance of repairing stalled replication forks. Nature 2000, 404, 37–41. [Google Scholar] [CrossRef]
- Michel, B.; Sinha, A.K.; Leach, D.R.F. Replication Fork Breakage and Restart in Escherichia coli. Microbiol. Mol. Biol. Rev. 2018, 82, e00013-18. [Google Scholar] [CrossRef]
- Cox, M.M. Relating biochemistry to biology: How the recombinational repair function of RecA protein is manifested in its molecular properties. BioEssays 1993, 15, 617–623. [Google Scholar] [CrossRef]
- Kreuzer, K.N. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 2005, 59, 43–67. [Google Scholar] [CrossRef]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 2018, 71, 135–147. [Google Scholar] [CrossRef]
- Spies, J.; Polasek-Sedlackova, H.; Lukas, J.; Somyajit, K. Homologous Recombination as a Fundamental Genome Surveillance Mechanism during DNA Replication. Genes 2021, 12, 1960. [Google Scholar] [CrossRef]
- Mosig, G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 1998, 32, 379–413. [Google Scholar] [CrossRef]
- Skalka, A. A Replicator’s View of Recombination (and Repair). In Mechanisms in Recombination; Grell, R.F., Ed.; Springer: Boston, MA, USA, 1974; pp. 421–432. [Google Scholar] [CrossRef]
- Moss, B. Origin of the poxviral membrane: A 50-year-old riddle. PLoS Pathog. 2018, 14, e1007002. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef]
- Moss, B. Poxviridae. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 2129–2159. [Google Scholar]
- Walsh, D. Poxviruses: Slipping and sliding through transcription and translation. PLoS Pathog. 2017, 13, e1006634. [Google Scholar] [CrossRef]
- Czarnecki, M.W.; Traktman, P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res. 2017, 234, 193–206. [Google Scholar] [CrossRef]
- Schmidt, F.I.; Bleck, C.K.E.; Mercer, J. Poxvirus host cell entry. Curr. Opin. Virol. 2012, 2, 20–27. [Google Scholar] [CrossRef]
- Roberts, K.L.; Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008, 16, 472–479. [Google Scholar] [CrossRef]
- Condit, R.C.; Moussatche, N.; Traktman, P. In A Nutshell: Structure and Assembly of the Vaccinia Virion. Adv. Virus Res. 2006, 66, 31–124. [Google Scholar] [CrossRef]
- Traktman, P. The Enzymology of Poxvirus DNA Replication. Curr. Top. Microbiol. Immunol. 1990, 163, 93–123. [Google Scholar] [CrossRef]
- Greseth, M.D.; Traktman, P. The Life Cycle of the Vaccinia Virus Genome. Annu. Rev. Virol. 2022, 9. [Google Scholar] [CrossRef]
- Tolonen, N.; Doglio, L.; Schleich, S.; Locker, J.K. Vaccinia Virus DNA Replication Occurs in Endoplasmic Reticulum-enclosed Cytoplasmic Mini-Nuclei. Mol. Biol. Cell 2001, 12, 2031–2046. [Google Scholar] [CrossRef]
- Cairns, J. The initiation of vaccinia infection. Virology 1960, 11, 603–623. [Google Scholar] [CrossRef]
- Paszkowski, P.; Noyce, R.S.; Evans, D.H. Live-Cell Imaging of Vaccinia Virus Recombination. PLoS Pathog. 2016, 12, e1005824. [Google Scholar] [CrossRef]
- Scutts, S.R.; Ember, S.W.; Ren, H.; Ye, C.; Lovejoy, C.A.; Mazzon, M.; Veyer, D.L.; Sumner, R.P.; Smith, G.L. DNA-PK Is Targeted by Multiple Vaccinia Virus Proteins to Inhibit DNA Sensing. Cell Rep. 2018, 25, 1953–1965.e4. [Google Scholar] [CrossRef]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266. [Google Scholar] [CrossRef]
- Kerr, S.M.; Johnston, L.H.; Odell, M.; Duncan, S.A.; Law, K.M.; Smith, G.L. Vaccinia DNA ligase complements Saccharomyces cerevisiae cdc9, localizes in cytoplasmic factories and affects virulence and virus sensitivity to DNA damaging agents. EMBO J. 1991, 10, 4343–4350. [Google Scholar] [CrossRef]
- Lin, Y.-C.J.; Li, J.; Irwin, C.R.; Jenkins, H.; DeLange, L.; Evans, D.H. Vaccinia Virus DNA Ligase Recruits Cellular Topoisomerase II to Sites of Viral Replication and Assembly. J. Virol. 2008, 82, 5922–5932. [Google Scholar] [CrossRef]
- Prescott, D.; Kates, J.; Kirkpatrick, J. Replication of vaccinia virus DNA in enucleated L-cells. J. Mol. Biol. 1971, 59, 505–508. [Google Scholar] [CrossRef]
- Jenner, E. An Inquiry into the Causes and Effects of the Variolæ Vaccinae, A Disease Discovered in Some of the Western Counties of England, Particularly Gloucestershire, and Known by the Name of the Cow Pox; Sampson Low: London, UK, 1798. [Google Scholar]
- Brinkmann, A.; Souza, A.R.V.; Esparza, J.; Nitsche, A.; Damaso, C.R. Re-assembly of nineteenth-century smallpox vaccine genomes reveals the contemporaneous use of horsepox and horsepox-related viruses in the USA. Genome Biol. 2020, 21, 1–6. [Google Scholar] [CrossRef]
- Tulman, E.R.; Delhon, G.; Afonso, C.L.; Lu, Z.; Zsak, L.; Sandybaev, N.T.; Kerembekova, U.Z.; Zaitsev, V.L.; Kutish, G.F.; Rock, D.L. Genome of Horsepox Virus. J. Virol. 2006, 80, 9244–9258. [Google Scholar] [CrossRef]
- Damaso, C.R. Revisiting Jenner’s mysteries, the role of the Beaugency lymph in the evolutionary path of ancient smallpox vaccines. Lancet Infect. Dis. 2018, 18, e55–e63. [Google Scholar] [CrossRef]
- Baxby, D. The Origins of Vaccinia Virus. J. Infect. Dis. 1977, 136, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Berry, G.P.; Dedrick, H.M. Method for changing the virus of rabbit fibroma (Shope) into that of infectious myxomatosis (Sanarelli). J. Bacteriol. 1936, 31, 50. [Google Scholar]
- Fenner, F.; Holmes, I.H.; Joklik, W.K.; Woodroofe, G.M.; Fenner, I.H.H.F. Reactivation of Heat-inactivated Poxviruses: A General Phenomenon which includes the Fibroma–Myxoma Virus Transformation of Berry and Dedrick. Nature 1959, 183, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Fenner, F.; Comben, B.M. Genetic studies with mammalian poxviruses: I. Demonstration of recombination between two strains of vaccinia virus. Virology 1958, 5, 530–548. [Google Scholar] [CrossRef]
- Gemmell, A.; Fenner, F. Genetic studies with mammalian poxviruses. III. White (u) mutants of rabbitpox virus. Virology 1960, 11, 219–235. [Google Scholar] [CrossRef]
- Gemmell, A.; Cairns, J. Linkage in the genome of an animal virus. Virology 1959, 8, 381–383. [Google Scholar] [CrossRef]
- Woodroofe, G.M.; Fenner, F. Genetic studies with mammalian poxviruses. IV. Hybridization between several different poxviruses. Virology 1960, 12, 272–282. [Google Scholar] [CrossRef]
- Strayer, D.S.; Skaletsky, E.; Cabirac, G.F.; Sharp, P.A.; Corbeil, L.B.; Sell, S.; Leibowitz, J.L. Malignant rabbit fibroma virus causes secondary immunosuppression in rabbits. J. Immunol. 1983, 130, 399–404. [Google Scholar]
- Upton, C.; Macen, J.; Maranchuk, R.; Delange, A.; McFadden, G. Tumorigenic poxviruses: Fine analysis of the recombination junctions in malignant rabbit fibroma virus, a recombinant between shope fibroma virus and myxoma virus. Virology 1988, 166, 229–239. [Google Scholar] [CrossRef]
- Bedson, H.S.; Dumbell, K.R. Hybrids derived from the viruses of variola major and cowpox. J. Hyg. 1964, 62, 147–158. [Google Scholar] [CrossRef]
- Bedson, H.S.; Dumbell, K.R. Hybrids derived from the viruses of alastrim and rabbit pox. J. Hyg. 1964, 62, 141–146. [Google Scholar] [CrossRef]
- Dumbell, K.R.; Bedson, H.S. The use of ceiling temperature and reactivation in the isolation of pox virus hybrids. J. Hyg. 1964, 62, 133–140. [Google Scholar] [CrossRef]
- Joklik, W.K. The poxviruses. Annu. Rev. Microbiol. 1968, 22, 359–390. [Google Scholar] [CrossRef]
- Stahl, F.W.; Edgar, R.S.; Steinberg, J. The Linkage Map of Bacteriophage T4. Genetics 1964, 50, 539–552. [Google Scholar] [CrossRef]
- Basilico, C.; Joklik, W.K. Studies on a temperature-sensitive mutant of vaccinia virus strain WR. Virology 1968, 36, 668–677. [Google Scholar] [CrossRef]
- Fenner, F.; Sambrook, J.F. Conditional lethal mutants of rabbitpox virus. II. Mutants (p) that fail to multiply in PK-2a cells. Virology 1966, 28, 600–609. [Google Scholar] [CrossRef]
- McClain, M.E.; Greenland, R.M. Recombination between rabbitpox virus mutants in permissive and nonpermissive cells. Virology 1965, 25, 516–522. [Google Scholar] [CrossRef]
- Magee, W.E.; Miller, O.V. Immunological evidence for the appearance of a new DNA-polymerase in cells infected with vaccinia virus. Virology 1967, 31, 64–69. [Google Scholar] [CrossRef]
- Berns, K.I.; Silverman, C.; Weissbach, A. Separation of a New Deoxyribonucleic Acid Polymerase from Vaccinia-infected HeLa Cells. J. Virol. 1969, 4, 15–23. [Google Scholar] [CrossRef]
- Jungwirth, C.; Joklik, W.K. Studies on “early” enzymes in HeLa cells infected with vaccinia virus. Virology 1965, 27, 80–93. [Google Scholar] [CrossRef]
- Citarella, R.V.; Muller, R.; Schlabach, A.; Weissbach, A. Studies on vaccinia virus-directed deoxyribonu-cleic acid polymerase. J. Virol. 1972, 10, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Dales, S.; Siminovitch, L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 1961, 10, 475–503. [Google Scholar] [CrossRef] [PubMed]
- Dales, S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J. Cell Biol. 1963, 18, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Dubbs, D.R.; Kit, S. Isolation and Properties of Vaccinia Mutants Deficient in Thymidine Ki-nase-Inducing Activity. Viroilogy 1964, 22, 214–225. [Google Scholar] [CrossRef]
- Kates, J.R.; McAuslan, B.R. Poxvirus DNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 1967, 58, 134–141. [Google Scholar] [CrossRef]
- Paoletti, E.; Moss, B. Deoxyribonucleic acid-dependent nucleotide phosphohydrolase activity in purified vaccinia virus. J. Virol. 1972, 10, 866–868. [Google Scholar] [CrossRef]
- Sambrook, J.; Shatkin, A.J. Polynucleotide ligase activity in cells infected with simian virus 40, polyoma virus, or vaccinia virus. J. Virol. 1969, 4, 719–726. [Google Scholar] [CrossRef]
- Oda, K.I.; Joklik, W.K. Hybridization and sedimentation studies on “early” and “late” vaccinia messenger RNA. J. Mol. Biol. 1967, 27, 395–419. [Google Scholar] [CrossRef]
- Moss, B.; Salzman, N.P. Sequential protein synthesis following vaccinia virus infection. J. Virol. 1968, 2, 1016–1027. [Google Scholar] [CrossRef]
- Moss, B. Regulation of vaccinia virus transcription. Annu. Rev. Biochem. 1990, 59, 661–688. [Google Scholar] [CrossRef]
- Sarov, I.; Becker, Y. Studies on vaccinia virus DNA. Virology 1967, 33, 369–375. [Google Scholar] [CrossRef]
- Geshelin, P.; Berns, K. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J. Mol. Biol. 1974, 88, 785–796. [Google Scholar] [CrossRef]
- Polisky, B.; Kates, J. Interaction of vaccinia DNA-binding proteins with DNA in vitro. Virology 1976, 69, 143–147. [Google Scholar] [CrossRef]
- Dahl, R.; Kates, J. Intracellular structures containing vaccinia DNA: Isolation and characterization. Virology 1970, 42, 453–462. [Google Scholar] [CrossRef]
- Soloski, M.J.; Esteban, M.; Holowczak, J.A. DNA-binding proteins in the cytoplasm of vaccinia virus-infected mouse L-cells. J. Virol. 1978, 25, 263–273. [Google Scholar] [CrossRef]
- Wittek, R.; Menna, A.; Schümperli, D.; Stoffel, S.; Müller, H.K.; Wyler, R. HindIII and Sst I restriction sites mapped on rabbit poxvirus and vaccinia virus DNA. J. Virol. 1977, 23, 669–678. [Google Scholar] [CrossRef]
- Fidzianska, E. Temperature mutants and recombinants of vaccinia virus. II. Recombination between temperature mutants. Acta Virol. 1972, 16, 298–307. [Google Scholar]
- Chernos, V.I.; Belanov, E.F.; Vasilieva, N.N. Temperature-sensitive mutants of vaccinia virus. I. Isolation and preliminary characterization. Acta Virol. 1978, 22, 81–90. [Google Scholar]
- Katz, E.; Margalith, E.; Winer, B. Genetic recombination between a temperature sensitive mutant and an isating beta thiosemicarbazone (IBT) resistant mutant of vaccinia virus. J. Antimicrob. Chemother. 1978, 4, 159–162. [Google Scholar] [CrossRef]
- Padgett, B.L.; Tomkins, J.K. Conditional lethal mutants of rabbitpox virus. 3. Temperature-sensitive (ts) mutants; physiological properties, complementation and recombination. Virology 1968, 36, 161–167. [Google Scholar] [CrossRef]
- Lake, J.R.; Silver, M.; Dales, S. Biogenesis of vaccinia: Complementation and recombination analysis of one group of conditional-lethal mutants defective in envelope self-assembly. Virology 1979, 96, 9–20. [Google Scholar] [CrossRef]
- Lackner, C.A.; D’Costa, S.M.; Buckb, C.; Condit, R.C. Complementation Analysis of the Dales Collection of Vaccinia Virus Temperature-Sensitive Mutants. Virology 2003, 305, 240–259. [Google Scholar] [CrossRef][Green Version]
- Tartaglia, J.; Paoletti, E. Physical mapping and DNA sequence analysis of the rifampicin resistance locus in vaccinia virus. Virology 1985, 147, 394–404. [Google Scholar] [CrossRef]
- McNulty-Kowalczyk, A.; Paoletti, E. Mutations in ORF D13L and Other Genetic Loci Alter the Rifampicin Phenotype of Vaccinia Virus. Virology 1993, 194, 638–646. [Google Scholar] [CrossRef]
- Ensinger, M.J. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus WR. J. Virol. 1982, 43, 778–790. [Google Scholar] [CrossRef]
- Ensinger, M.J.; Rovinsky, M. Marker rescue of temperature-sensitive mutations of vaccinia virus WR: Correlation of genetic and physical maps. J. Virol. 1983, 48, 419–428. [Google Scholar] [CrossRef]
- Ensinger, M.J.; Weir, J.P.; Moss, B. Fine structure marker rescue of temperature-sensitive mutations of vaccinia virus within a central conserved region of the genome. J. Virol. 1985, 56, 1027–1029. [Google Scholar] [CrossRef]
- Kato, S.E.; Moussatche, N.; D’Costa, S.M.; Bainbridge, T.W.; Prins, C.; Strahl, A.L.; Shatzer, A.N.; Brinker, A.J.; Kay, N.E.; Condit, R.C. Marker rescue mapping of the combined Condit/Dales collection of tempera-ture-sensitive vaccinia virus mutants. Virology 2008, 375, 213–222. [Google Scholar] [CrossRef][Green Version]
- Seto, J.; Celenza, L.M.; Condit, R.C.; Niles, E.G. Genetic map of the vaccinia virus HindIII D Fragment. Virology 1987, 160, 110–119. [Google Scholar] [CrossRef]
- Fathi, Z.; Dyster, L.M.; Seto, J.; Condit, R.C.; Niles, E.G. Intragenic and intergenic recombination between temperature-sensitive mutants of vaccinia virus. J. Gen. Virol. 1991, 72, 2733–2737. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.J.; Evans, D.H. Vaccinia Virus Particles Mix Inefficiently, and in a Way That Would Restrict Viral Recombination, in Coinfected Cells. J. Virol. 2010, 84, 2432–2443. [Google Scholar] [CrossRef]
- Fenner, F.; Henderson, A.; Arita, I.; Jezek, Z.; Ladnyi, I.D. Smallpox and Its Eradication; World Health Organization: Geneva, Switzerland, 1988. [Google Scholar]
- Behbehani, A.M. The smallpox story: Life and death of an old disease. Microbiol. Rev. 1983, 47, 455–509. [Google Scholar] [CrossRef]
- Graham, F.L.; Van Der Eb, A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973, 52, 456–467. [Google Scholar] [CrossRef]
- Sam, C.K.; Dumbell, K.R. Expression of Poxvirus DNA in Co-Infected Cells and Marker Rescue of Thermosensitive Mutants by Subgenomic Fragments of DNA. Ann. Virol. 1981, 132, 135–150. [Google Scholar]
- Weir, J.P.; Bajszár, G.; Moss, B. Mapping of the vaccinia virus thymidine kinase gene by marker rescue and by cell-free translation of selected mRNA. Proc. Natl. Acad. Sci. USA 1982, 79, 1210–1214. [Google Scholar] [CrossRef]
- Nakano, E.; Panicali, D.; Paoletti, E. Molecular genetics of vaccinia virus: Demonstration of marker rescue. Proc. Natl. Acad. Sci. USA 1982, 79, 1593–1596. [Google Scholar] [CrossRef]
- DeLange, A.M.; McFadden, G. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc. Natl. Acad. Sci. USA 1986, 83, 614–618. [Google Scholar] [CrossRef]
- Panicali, D.; Paoletti, E. Construction of poxviruses as cloning vectors: Insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc. Natl. Acad. Sci. USA 1982, 79, 4927–4931. [Google Scholar] [CrossRef]
- Matía, A.; Lorenzo, M.M.; Blasco, R. Tools for the targeted genetic modification of poxvirus genomes. Curr. Opin. Virol. 2020, 44, 183–190. [Google Scholar] [CrossRef]
- Volz, A.; Sutter, G. Modified Vaccinia Virus Ankara: History, Value in Basic Research, and Current Perspectives for Vaccine Development. Adv. Virus Res. 2017, 97, 187–243. [Google Scholar]
- Sánchez-Sampedro, L.; Perdiguero, B.; Mejías-Pérez, E.; García-Arriaza, J.; Di Pilato, M.; Esteban, M. The Evolution of Poxvirus Vaccines. Viruses 2015, 7, 1726–1803. [Google Scholar] [CrossRef]
- Moss, B. Reflections on the early development of poxvirus vectors. Vaccine 2013, 31, 4220–4222. [Google Scholar] [CrossRef]
- Kirn, D.H.; Thorne, S.H. Targeted and armed oncolytic poxviruses: A novel multi-mechanistic therapeutic class for cancer. Nat. Cancer 2009, 9, 64–71. [Google Scholar] [CrossRef]
- Paoletti, E. Applications of pox virus vectors to vaccination: An update. Proc. Natl. Acad. Sci. USA 1996, 93, 11349–11353. [Google Scholar] [CrossRef]
- Wyatt, L.S.; Earl, P.L.; Moss, B. Generation of Recombinant Vaccinia Viruses. Curr. Protoc. Mol. Biol. 2017, 117, 16.17.1–16.17.18. [Google Scholar] [CrossRef]
- Condit, R.C.; Motyczka, A.; Spizz, G. Isolation, characterization, and physical mapping of tempera-ture-sensitive mutants of vaccinia virus. Virology 1983, 128, 429–443. [Google Scholar] [CrossRef]
- Drillen, R.; Spehner, D. Physical mapping of vaccinia virus temperature-sensitive mutations. Virology 1983, 131, 385–393. [Google Scholar] [CrossRef]
- Traktman, P.; Kelvin, M.; Pacheco, S. Molecular genetic analysis of vaccinia virus DNA polymerase mutants. J. Virol. 1989, 63, 841–846. [Google Scholar] [CrossRef]
- Condit, R.C.; Niles, E.G. Orthopoxvirus genetics. Curr. Top. Microbiol. Immunol. 1990, 163, 1–39. [Google Scholar]
- Liu, R.; Mendez-Rios, J.D.; Peng, C.; Xiao, W.; Weisberg, A.S.; Wyatt, L.S.; Moss, B. SPI-1 is a missing host-range factor required for replication of the attenuated modified vaccinia Ankara (MVA) vaccine vector in human cells. PLoS Pathog. 2019, 15, e1007710. [Google Scholar] [CrossRef]
- Taddie, J.A.; Traktman, P. Genetic characterization of the vaccinia virus DNA polymerase: Identifica-tion of point mutations conferring altered drug sensitivities and reduced fidelity. J. Virol. 1991, 65, 869–879. [Google Scholar] [CrossRef] [PubMed]
- DeLange, A.M.; Carpenter, M.S.; Choy, J.; Newsway, V.E. An etoposide-induced block in vaccinia virus telomere resolution is dependent on the virus-encoded DNA ligase. J. Virol. 1995, 69, 2082–2091. [Google Scholar] [CrossRef] [PubMed]
- Andrei, G.; Gammon, D.B.; Fiten, P.; De Clercq, E.; Opdenakker, G.; Snoeck, R.; Evans, D.H. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 2006, 80, 9391–9401. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.A. High-frequency homologous recombination in vaccinia virus DNA. J. Virol. 1987, 61, 1788–1795. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Brechling, K.; Moss, B. Vaccinia virus expression vector: Coexpression of be-ta-galactosidase provides visual screening of recombinant virus plaques. Mol. Cell. Biol. 1985, 5, 3403–3409. [Google Scholar]
- Boyle, D.B.; Coupar, B.E. A dominant selectable marker for the construction of recombinant poxviruses. Gene 1988, 65, 123–128. [Google Scholar] [CrossRef]
- Domınguez, J.; Lorenzo, M.d.M.; Blasco, R. Green fluorescent protein expressed by a recombinant vaccinia virus permits early detection of infected cells by flow cytometry. J. Immunol. Methods 1998, 220, 115–121. [Google Scholar] [CrossRef]
- Dower, K.; Rubins, K.H.; Hensley, L.E.; Connor, J.H. Development of Vaccinia reporter viruses for rapid, high content analysis of viral function at all stages of gene expression. Antivir. Res. 2011, 91, 72–80. [Google Scholar] [CrossRef]
- Rintoul, J.L.; Wang, J.; Gammon, D.B.; Van Buuren, N.J.; Garson, K.; Jardine, K.; Barry, M.; Evans, D.H.; Bell, J.C. A Selectable and Excisable Marker System for the Rapid Creation of Recombinant Poxviruses. PLoS ONE 2011, 6, e24643. [Google Scholar] [CrossRef]
- Weller, S.K.; Sawitzke, J.A. Recombination promoted by DNA viruses: Phage lambda to herpes simplex virus. Annu. Rev. Microbiol. 2014, 68, 237–258. [Google Scholar] [CrossRef]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef]
- West, S.C.; Kowalczykowski, S.C.; Charles, M. Radding: A love of science and art. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Benson, F.E.; Stasiak, A.; West, S.C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994, 13, 5764–5771. [Google Scholar] [CrossRef]
- Szostak, J.W.; Orr-Weaver, T.L.; Rothstein, R.J.; Stahl, F.W. The double-strand-break repair model for recombination. Cell 1983, 33, 25–35. [Google Scholar] [CrossRef]
- Zhang, W.; Evans, D.H. DNA strand exchange catalyzed by proteins from vaccinia virus-infected cells. J. Virol. 1993, 67, 204–212. [Google Scholar] [CrossRef]
- Willer, D.O.; Mann, M.J.; Zhang, W.; Evans, D.H. Vaccinia Virus DNA Polymerase Promotes DNA Pairing and Strand-Transfer Reactions. Virology 1999, 257, 511–523. [Google Scholar] [CrossRef][Green Version]
- Brewster, J.L.; Tolun, G. Half a century of bacteriophage lambda recombinase: In vitro studies of lambda exonuclease and Red-beta annealase. IUBMB Life 2020, 72, 1622–1633. [Google Scholar] [CrossRef]
- Reuven, N.B.; Staire, A.E.; Myers, R.S.; Weller, S.K. The Herpes Simplex Virus Type 1 Alkaline Nuclease and Single-Stranded DNA Binding Protein Mediate Strand Exchange In Vitro. J. Virol. 2003, 77, 7425–7433. [Google Scholar] [CrossRef]
- Stahl, M.M.; Thomason, L.; Poteete, A.R.; Tarkowski, T.; Kuzminov, A.; Stahl, F.W. Annealing vs. invasion in phage lambda recombination. Genetics 1997, 147, 961–977. [Google Scholar] [CrossRef]
- Schumacher, A.J.; Mohni, K.N.; Kan, Y.; Hendrickson, E.A.; Stark, J.M.; Weller, S.K. The HSV-1 Exonuclease, UL12, Stimulates Recombination by a Single Strand Annealing Mechanism. PLoS Pathog. 2012, 8, e1002862. [Google Scholar] [CrossRef]
- Odom, M.R.; Hendrickson, R.C.; Lefkowitz, E.J. Poxvirus protein evolution: Family wide assessment of possible horizontal gene transfer events. Virus Res. 2009, 144, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Hertig, C.; Coupar, B.E.; Gould, A.R.; Boyle, D.B. Field and Vaccine Strains of Fowlpox Virus Carry Integrated Sequences from the Avian Retrovirus, Reticuloendotheliosis Virus. Virology 1997, 235, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Gubser, C.; Bergamaschi, D.; Hollinshead, M.; Lu, X.; Van Kuppeveld, F.J.M.; Smith, G.L. A New Inhibitor of Apoptosis from Vaccinia Virus and Eukaryotes. PLoS Pathog. 2007, 3, e17. [Google Scholar] [CrossRef] [PubMed]
- Garcel, A.; Crance, J.M.; Drillien, R.; Garin, D.; Favier, A.L. Genomic sequence of a clonal isolate of the vaccinia virus Lister strain employed for smallpox vaccination in France and its comparison to other orthopoxviruses. J. Gen. Virol. 2007, 88, 1906–1916. [Google Scholar] [CrossRef]
- Fixsen, S.M.; Cone, K.R.; Goldstein, S.A.; Sasani, T.A.; Quinlan, A.R.; Rothenburg, S.; Elde, N.C. Poxviruses capture host genes by LINE-1 retrotransposition. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rahman, M.J.; Haller, S.L.; Stoian, A.M.M.; Li, J.; Brennan, G.; Rothenburg, S. LINE-1 retrotransposons facilitate horizontal gene transfer into poxviruses (preprint). bioRxiv 2021. [Google Scholar] [CrossRef]
- Vallée, G.; Norris, P.; Paszkowski, P.; Noyce, R.S.; Evans, D.H. Vaccinia Virus Gene Acquisition through Nonhomologous Recombination. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Luteijn, R.D.; Drexler, I.; Smith, G.L.; Lebbink, R.J.; Wiertz, E.J.H.J. Mutagenic repair of double-stranded DNA breaks in vaccinia virus genomes requires cellular DNA ligase IV activity in the cytosol. J. Gen. Virol. 2018, 99, 790–804. [Google Scholar] [CrossRef]
- Evans, D.H.; Stuart, D.; McFadden, G. High levels of genetic recombination among cotransfected plasmid DNAs in poxvirus-infected mammalian cells. J. Virol. 1988, 62, 367–375. [Google Scholar] [CrossRef]
- Slabaugh, M.; Roseman, N.; Mathews, C. Amplification of the ribonucleotide reductase small subunit gene: Analysis of novel joints and the mechanism of gene duplication in vaccinia virus. Nucleic Acids Res. 1989, 17, 7073–7088. [Google Scholar] [CrossRef]
- Baroudy, B.M.; Moss, B. Sequence homologies of diverse length tandem repetitions near ends of vaccinia virus genome suggest unequal crossing over. Nucleic Acids Res. 1982, 10, 5673–5679. [Google Scholar] [CrossRef]
- Coulson, D.; Upton, C. Characterization of indels in poxvirus genomes. Virus Genes 2010, 42, 171–177. [Google Scholar] [CrossRef]
- Parks, R.J.; Evans, D.H. Effect of marker distance and orientation on recombinant formation in poxvirus-infected cells. J. Virol. 1991, 65, 1263–1272. [Google Scholar] [CrossRef]
- Stahl, F.W. Genetic Recombination: Thinking about It in Phage and Fungi; W. H. Freeman: San Francisco, CA, USA, 1979. [Google Scholar]
- Fisher, C.; Parks, R.J.; Lauzon, M.L.; Evans, D. Heteroduplex DNA formation is associated with replication and recombination in poxvirus-infected cells. Genetics 1991, 129, 7–18. [Google Scholar] [CrossRef]
- Bortner, C.; Hernandez, T.R.; Lehman, I.; Griffith, J. Herpes Simplex Virus 1 Single-strand DNA-binding Protein (ICP8) Will Promote Homologous Pairing and Strand Transfer. J. Mol. Biol. 1993, 231, 241–250. [Google Scholar] [CrossRef]
- Kolodner, R.; Evans, D.H.; Morrison, P.T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc. Natl. Acad. Sci. USA 1987, 84, 5560–5564. [Google Scholar] [CrossRef]
- Stanitsa, E.S.; Arps, L.; Traktman, P. Vaccinia Virus Uracil DNA Glycosylase Interacts with the A20 Protein to Form a Heterodimeric Processivity Factor for the Viral DNA Polymerase. J. Biol. Chem. 2006, 281, 3439–3451. [Google Scholar] [CrossRef]
- Mcdonald, W.; Traktman, P. Overexpression and Purification of the Vaccinia Virus DNA Polymerase. Protein Expr. Purif. 1994, 5, 409–421. [Google Scholar] [CrossRef]
- Rochester, S.C.; Traktman, P. Characterization of the Single-Stranded DNA Binding Protein Encoded by the Vaccinia Virus I3 Gene. J. Virol. 1998, 72. [Google Scholar] [CrossRef]
- Tseng, M.; Palaniyar, N.; Zhang, W.; Evans, D.H. DNA Binding and Aggregation Properties of the Vaccinia Virus I3L Gene Product. J. Biol. Chem. 1999, 274, 21637–21644. [Google Scholar] [CrossRef]
- Greseth, M.D.; Czarnecki, M.W.; Bluma, M.S.; Traktman, P. Isolation and Characterization of vDeltaI3 Confirm that Vaccinia Virus SSB Plays an Essential Role in Viral Replication. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Willer, D.O.; Yao, X.-D.; Mann, M.J.; Evans, D. In Vitro Concatemer Formation Catalyzed by Vaccinia Virus DNA Polymerase. Virology 2000, 278, 562–569. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef]
- Hamilton, M.D.; Evans, D.H. Enzymatic processing of replication and recombination intermediates by the vaccinia virus DNA polymerase. Nucleic Acids Res. 2005, 33, 2259–2268. [Google Scholar] [CrossRef][Green Version]
- Hamilton, M.D.; Nuara, A.A.; Gammon, D.B.; Buller, R.M.; Evans, D.H. Duplex strand joining reactions catalyzed by vaccinia virus DNA polymerase. Nucleic Acids Res. 2006, 35, 143–151. [Google Scholar] [CrossRef]
- Reha-Krantz, L.J.; Nonay, R.L. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3′-->5′-exonuclease activity. J. Biol. Chem. 1993, 268, 27100–27108. [Google Scholar] [CrossRef]
- Gammon, D.B.; Evans, D.H. The 3′-to-5′ Exonuclease Activity of Vaccinia Virus DNA Polymerase Is Essential and Plays a Role in Promoting Virus Genetic Recombination. J. Virol. 2009, 83, 4236–4250. [Google Scholar] [CrossRef]
- Magee, W.C.; Hostetler, K.; Evans, D.H. Mechanism of Inhibition of Vaccinia Virus DNA Polymerase by Cidofovir Diphosphate. Antimicrob. Agents Chemother. 2005, 49, 3153–3162. [Google Scholar] [CrossRef]
- Yao, X.-D.; Evans, D.H. Effects of DNA Structure and Homology Length on Vaccinia Virus Recombination. J. Virol. 2001, 75, 6923–6932. [Google Scholar] [CrossRef]
- Yao, X.-D.; Evans, D.H. Characterization of the recombinant joints formed by single-strand annealing reactions in vaccinia virus-infected cells. Virology 2003, 308, 147–156. [Google Scholar] [CrossRef][Green Version]
- McFadden, G.; Morgan, A.R. DNA cruciform structures: Implications for telomer replication in eukaryotes and instability of long palindromic DNA sequences in prokaryotes. J. Theor. Biol. 1982, 97, 343–349. [Google Scholar] [CrossRef]
- Garcia, A.D.; Aravind, L.; Koonin, E.V.; Moss, B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl. Acad. Sci. USA 2000, 97, 8926–8931. [Google Scholar] [CrossRef] [PubMed]
- Culyba, M.J.; Minkah, N.; Hwang, Y.; Benhamou, O.-M.J.; Bushman, F.D. DNA Branch Nuclease Activity of Vaccinia A22 Resolvase. J. Biol. Chem. 2007, 282, 34644–34652. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.; Shen, L.; Tcherepanov, V.; Watson, C.; Upton, C. Predicted function of the vaccinia virus G5R protein. Bioinformatics 2006, 22, 2846–2850. [Google Scholar] [CrossRef][Green Version]
- Senkevich, T.G.; Koonin, E.V.; Moss, B. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc. Natl. Acad. Sci. USA 2009, 106, 17921–17926. [Google Scholar] [CrossRef]
- Shuman, S. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 1994, 269, 32678–32684. [Google Scholar] [CrossRef]
- Shuman, S.; Prescott, J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 1990, 265, 17826–17836. [Google Scholar] [CrossRef]
- Palaniyar, N.; Gerasimopoulos, E.; Evans, D. Shope fibroma virus DNA topoisomerase catalyses holliday junction resolution and hairpin formation in vitro. J. Mol. Biol. 1999, 287, 9–20. [Google Scholar] [CrossRef]
- Palaniyar, N.; Fisher, C.; Parks, R.; Evans, D.H. SFV Topoisomerase: Sequence Specificity in a Genetically Mapped Interval. Virology 1996, 221, 351–354. [Google Scholar] [CrossRef][Green Version]
- Kerr, S.M.; Smith, G.L. Vaccinia virus encodes a polypeptide with DNA ligase activity. Nucleic Acids Res. 1989, 17, 9039–9050. [Google Scholar] [CrossRef]
- Evans, E.; Klemperer, N.; Ghosh, R.; Traktman, P. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 1995, 69, 5353–5361. [Google Scholar] [CrossRef]
- Kilcher, S.; Schmidt, F.I.; Schneider, C.; Kopf, M.; Helenius, A.; Mercer, J. siRNA Screen of Early Poxvirus Genes Identifies the AAA+ ATPase D5 as the Virus Genome-Uncoating Factor. Cell Host Microbe 2014, 15, 103–112. [Google Scholar] [CrossRef]
- Hutin, S.; Ling, W.L.; Round, A.; Effantin, G.; Reich, S.; Iseni, F.; Tarbouriech, N.; Schoehn, G.; Burmeister, W.P. Domain Organization of Vaccinia Virus Helicase-Primase D5. J. Virol. 2016, 90, 4604–4613. [Google Scholar] [CrossRef]
- De Silva, F.S.; Lewis, W.; Berglund, P.; Koonin, E.V.; Moss, B. Poxvirus DNA primase. Proc. Natl. Acad. Sci. USA 2007, 104, 18724–18729. [Google Scholar] [CrossRef]
- Upton, C.; Stuart, D.T.; McFadden, G. Identification of a poxvirus gene encoding a uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA 1993, 90, 4518–4522. [Google Scholar] [CrossRef]
- Tarbouriech, N.; Ducournau, C.; Hutin, S.; Mas, P.J.; Man, P.; Forest, E.; Hart, D.J.; Peyrefitte, C.N.; Burmeister, W.P.; Iseni, F. The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Klemperer, N.; McDonald, W.; Boyle, K.; Unger, B.; Traktman, P. The A20R Protein Is a Stoichiometric Component of the Processive Form of Vaccinia Virus DNA Polymerase. J. Virol. 2001, 75, 12298–12307. [Google Scholar] [CrossRef]
- McFadden, G.; Dales, S. Biogenesis of poxviruses: Mirror-image deletions in vaccinia virus DNA. Cell 1979, 18, 101–108. [Google Scholar] [CrossRef]
- Sprygin, A.; Babin, Y.; Pestova, Y.; Kononova, S.; Wallace, D.B.; Van Schalkwyk, A.; Byadovskaya, O.; Diev, V.; Lozovoy, D.; Kononov, A. Analysis and insights into recombination signals in lumpy skin disease virus recovered in the field. PLoS ONE 2018, 13, e0207480. [Google Scholar] [CrossRef]
- Gershon, P.D.; Kitching, R.P.; Hammond, J.M.; Black, D.N. Poxvirus Genetic Recombination during Natural Virus Transmission. J. Gen. Virol. 1989, 70, 485–489. [Google Scholar] [CrossRef]
- Biswas, S.; Noyce, R.S.; Babiuk, L.A.; Lung, O.; Bulach, D.M.; Bowden, T.R.; Boyle, D.B.; Babiuk, S.; Evans, D.H. Extended sequencing of vaccine and wild-type capripoxvirus isolates provides insights into genes modulating virulence and host range. Transbound. Emerg. Dis. 2019, 67, 80–97. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome Sequence Diversity and Clues to the Evolution of Variola (Smallpox) Virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Upton, C.; Hazes, B.; Evans, D.H. Genomic Analysis of the Vaccinia Virus Strain Variants Found in Dryvax Vaccine. J. Virol. 2011, 85, 13049–13060. [Google Scholar] [CrossRef]
- Esparza, J.; Lederman, S.; Nitsche, A.; Damaso, C.R. Early smallpox vaccine manufacturing in the United States: Introduction of the “animal vaccine” in 1870, establishment of “vaccine farms”, and the beginnings of the vaccine industry. Vaccine 2020, 38, 4773–4779. [Google Scholar] [CrossRef]
- Duggan, A.T.; Klunk, J.; Porter, A.; Dhody, A.N.; Hicks, R.; Smith, G.L.; Humphreys, M.; McCollum, A.M.; Davidson, W.B.; Wilkins, K.; et al. The origins and genomic diversity of American Civil War Era smallpox vaccine strains. Genome Biol. 2020, 21, 175. [Google Scholar] [CrossRef]
- Qin, L.; Evans, D.H. Genome Scale Patterns of Recombination between Coinfecting Vaccinia Viruses. J. Virol. 2014, 88, 5277–5286. [Google Scholar] [CrossRef]
- Tu, S.-L.; Staheli, J.P.; McClay, C.; McLeod, K.; Rose, T.M.; Upton, C. Base-By-Base Version 3: New Comparative Tools for Large Virus Genomes. Viruses 2018, 10, 637. [Google Scholar] [CrossRef]
- Willer, D.O.; McFadden, G.; Evans, D.H. The Complete Genome Sequence of Shope (Rabbit) Fibroma Virus. Virology 1999, 264, 319–343. [Google Scholar] [CrossRef]
- Elde, N.C.; Child, S.J.; Eickbush, M.T.; Kitzman, J.O.; Rogers, K.S.; Shendure, J.; Geballe, A.P.; Malik, H.S. Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses. Cell 2012, 150, 831–841. [Google Scholar] [CrossRef]
- Sasani, T.A.; Cone, K.R.; Quinlan, A.R.; Elde, N.C. Long read sequencing reveals poxvirus evolution through rapid homogenization of gene arrays. eLife 2018, 7. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Yutin, N.; Wolf, Y.I.; Koonin, E.V.; Moss, B. Ancient Gene Capture and Recent Gene Loss Shape the Evolution of Orthopoxvirus-Host Interaction Genes. mBio 2021, 12, e0149521. [Google Scholar] [CrossRef]
- Kieser, Q.; Paszkowski, P.; Lin, J.; Evans, D.; Noyce, R. Visualizing Poxvirus Replication and Recombination Using Live-Cell Imaging. Methods Mol. Biol. 2019, 2023, 221–235. [Google Scholar] [CrossRef]
- Kieser, Q.; Noyce, R.S.; Shenouda, M.; Lin, Y.-C.J.; Evans, D.H. Cytoplasmic factories, virus assembly, and DNA replication kinetics collectively constrain the formation of poxvirus recombinants. PLoS ONE 2020, 15, e0228028. [Google Scholar] [CrossRef]
- De Silva, F.S.; Moss, B. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Virol. J. 2005, 2, 23. [Google Scholar] [CrossRef]
- Smith, G.L.; Moss, B. Infectious poxvirus vectors have capacity for at least 25 000 base pairs of foreign DNA. Gene 1983, 25, 21–28. [Google Scholar] [CrossRef]
- Woolsey, C.; Geisbert, T.W. Current state of Ebola virus vaccines: A snapshot. PLoS Pathog. 2021, 17, e1010078. [Google Scholar] [CrossRef]
- Falkner, F.G.; Moss, B. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 1990, 64, 3108–3111. [Google Scholar] [CrossRef]
- Marzook, N.B.; Procter, D.J.; Lynn, H.; Yamamoto, Y.; Horsington, J.; Newsome, T.P. Methodology for the efficient generation of fluorescently tagged vaccinia virus proteins. J. Vis. Exp. 2014, e51151. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, W.; Wang, J.; Al Yaghchi, C.; Ahmed, J.; Chard, L.; Lemoine, N.R.; Wang, Y. Efficiently Editing the Vaccinia Virus Genome by Using the CRISPR-Cas9 System. J. Virol. 2015, 89, 5176–5179. [Google Scholar] [CrossRef]
- Gowripalan, A.; Smith, S.; Stefanovic, T.; Tscharke, D.C. Rapid poxvirus engineering using CRISPR/Cas9 as a selection tool. Commun. Biol. 2020, 3, 643. [Google Scholar] [CrossRef]
- Marsischky, G.; LaBaer, J. Many Paths to Many Clones: A Comparative Look at High-Throughput Cloning Methods. Genome Res. 2004, 14, 2020–2028. [Google Scholar] [CrossRef][Green Version]
- Irwin, C.R.; Farmer, A.; Willer, D.O.; Evans, D.H. In-fusion(R) cloning with vaccinia virus DNA polymerase. Methods Mol. Biol. 2012, 890, 23–35. [Google Scholar]
- Domi, A.; Moss, B. Engineering of a vaccinia virus bacterial artificial chromosome in Escherichia coli by bacteriophage lambda-based recombination. Nat. Methods 2005, 2, 95–97. [Google Scholar] [CrossRef]
- Meisinger-Henschel, C.; Späth, M.; Lukassen, S.; Wolferstätter, M.; Kachelriess, H.; Baur, K.; Dirmeier, U.; Wagner, M.; Chaplin, P.; Suter, M.; et al. Introduction of the Six Major Genomic Deletions of Modified Vaccinia Virus Ankara (MVA) into the Parental Vaccinia Virus Is Not Sufficient To Reproduce an MVA-Like Phenotype in Cell Culture and in Mice. J. Virol. 2010, 84, 9907–9919. [Google Scholar] [CrossRef]
- Peng, C.; Moss, B. Repair of a previously uncharacterized second host-range gene contributes to full replication of modified vaccinia virus Ankara (MVA) in human cells. Proc. Natl. Acad. Sci. USA 2020, 117, 3759–3767. [Google Scholar] [CrossRef]
- Yao, X.-D.; Evans, D.H. High-Frequency Genetic Recombination and Reactivation of Orthopoxviruses from DNA Fragments Transfected into Leporipoxvirus-Infected Cells. J. Virol. 2003, 77, 7281–7290. [Google Scholar] [CrossRef]
- Laudermilch, E.; Chandran, K. MAVERICC: Marker-free Vaccinia Virus Engineering of Recombinants through in vitro CRISPR/Cas9 Cleavage. J. Mol. Biol. 2021, 433, 166896. [Google Scholar] [CrossRef]
- Noyce, R.S.; Lederman, S.; Evans, D.H. Construction of an infectious horsepox virus vaccine from chemically synthesized DNA fragments. PLoS ONE 2018, 13, e0188453. [Google Scholar] [CrossRef]
- Chiuppesi, F.; Salazar, M.D.; Contreras, H.; Nguyen, V.; Martinez, J.; Park, S.; Nguyen, J.; Kha, M.; Iniguez, A.; Zhou, Q.; et al. Development of a Multi-Antigenic SARS-CoV-2 Vaccine Using a Synthetic Poxvirus Platform. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Shenouda, M.M.; Noyce, R.S.; Lee, S.Z.; Wang, J.L.; Lin, Y.-C.; Favis, N.A.; Desaulniers, M.A.; Evans, D.H. The mismatched nucleotides encoded in vaccinia virus flip-and-flop hairpin telomeres serve an essential role in virion maturation. PLoS Pathog. 2022, 18, e1010392. [Google Scholar] [CrossRef] [PubMed]
- Noyce, R.S.; Evans, D.H. Synthetic horsepox viruses and the continuing debate about dual use research. PLoS Pathog. 2018, 14, e1007025. [Google Scholar] [CrossRef] [PubMed]
- Merchlinsky, M. Intramolecular homologous recombination in cells infected with temperature-sensitive mutants of vaccinia virus. J. Virol. 1989, 63, 2030–2035. [Google Scholar] [CrossRef] [PubMed]
- Pogo, B.G. Elimination of naturally occurring crosslinks in vaccinia virus DNA after viral penetration into cells. Proc. Natl. Acad. Sci. USA 1977, 74, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Moyer, R.W.; Graves, R.L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell 1981, 27, 391–401. [Google Scholar] [CrossRef]
- Pogo, B.G.; Berkowitz, E.; Dales, S. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology 1984, 132, 436–444. [Google Scholar] [CrossRef]
- Parker, R.F. Studies of the Infectious Unit of Myxoma. J. Exp. Med. 1940, 71, 439–444. [Google Scholar] [CrossRef]
- Du, S.; Traktman, P. Vaccinia virus DNA replication: Two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc. Natl. Acad. Sci. USA 1996, 93, 9693–9698. [Google Scholar] [CrossRef]
- Willis, N.A.; Chandramouly, G.; Huang, B.; Kwok, A.; Follonier, C.; Deng, C.; Scully, R. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature 2014, 510, 556–559. [Google Scholar] [CrossRef]
- Marie, L.; Symington, L.S. Mechanism for inverted-repeat recombination induced by a replication fork barrier. Nat. Commun. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Larsen, N.B.; Sass, E.; Suski, C.; Mankouri, H.W.; Hickson, I.D. The Escherichia coli Tus–Ter replication fork barrier causes site-specific DNA replication perturbation in yeast. Nat. Commun. 2014, 5, 3574. [Google Scholar] [CrossRef]
- Hogg, M.; Aller, P.; Konigsberg, W.; Wallace, S.S.; Doublie, S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2007, 282, 1432–1444. [Google Scholar] [CrossRef]
- McCraith, S.; Holtzman, T.; Moss, B.; Fields, S. Genome-wide analysis of vaccinia virus protein–protein interactions. Proc. Natl. Acad. Sci. USA 2000, 97, 4879–4884. [Google Scholar] [CrossRef]
| Protein | Gene | Biochemical Activity | Role in Recombination | Reference |
|---|---|---|---|---|
| DNA polymerase | E9L | 3′-5′ exonuclease 5′-3′ polymerase | Strand transferase | [16,147,152] |
| Single-strand DNA binding protein | I3L | Replicative high affinity SSB | Stimulates E9-catalyzed strand transfer | [144,145,147] |
| RuvC-like Holliday junction resolvase | A22R | Branch-specific endonuclease | Cleaves 3- and 4-branched DNAs | [157,158] |
| FEN1-like flap endonuclease | G5R | Single-strand endonuclease | Cleaves single-stranded DNA flaps | [159,160] |
| DNA ligase | A48R | DNA ligase, binds cell topoisomerase II | Repairs nicks | [28,165] |
| ATPase-helicase and primase | D5R | Helicase (putative), uncoating, primase | (hypothetical) | [166,167,168,169] |
| Uracil glycosylase | D4R | Uracil glycosylase, E9 processivity component | (hypothetical) | [16,142,170,171] |
| A20 | A20R | E9 processivity factor | (hypothetical) | [16,171,172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evans, D.H. Poxvirus Recombination. Pathogens 2022, 11, 896. https://doi.org/10.3390/pathogens11080896
Evans DH. Poxvirus Recombination. Pathogens. 2022; 11(8):896. https://doi.org/10.3390/pathogens11080896
Chicago/Turabian StyleEvans, David Hugh. 2022. "Poxvirus Recombination" Pathogens 11, no. 8: 896. https://doi.org/10.3390/pathogens11080896
APA StyleEvans, D. H. (2022). Poxvirus Recombination. Pathogens, 11(8), 896. https://doi.org/10.3390/pathogens11080896






