Characterization of Bordetella pertussis Strains Isolated from India
Abstract
:1. Introduction
2. Results
2.1. Isolate Characterization
2.2. Fimbrial Serotyping
2.3. Genotypic Analysis
2.4. Multilocus Antigen Sequence Typing (MAST)
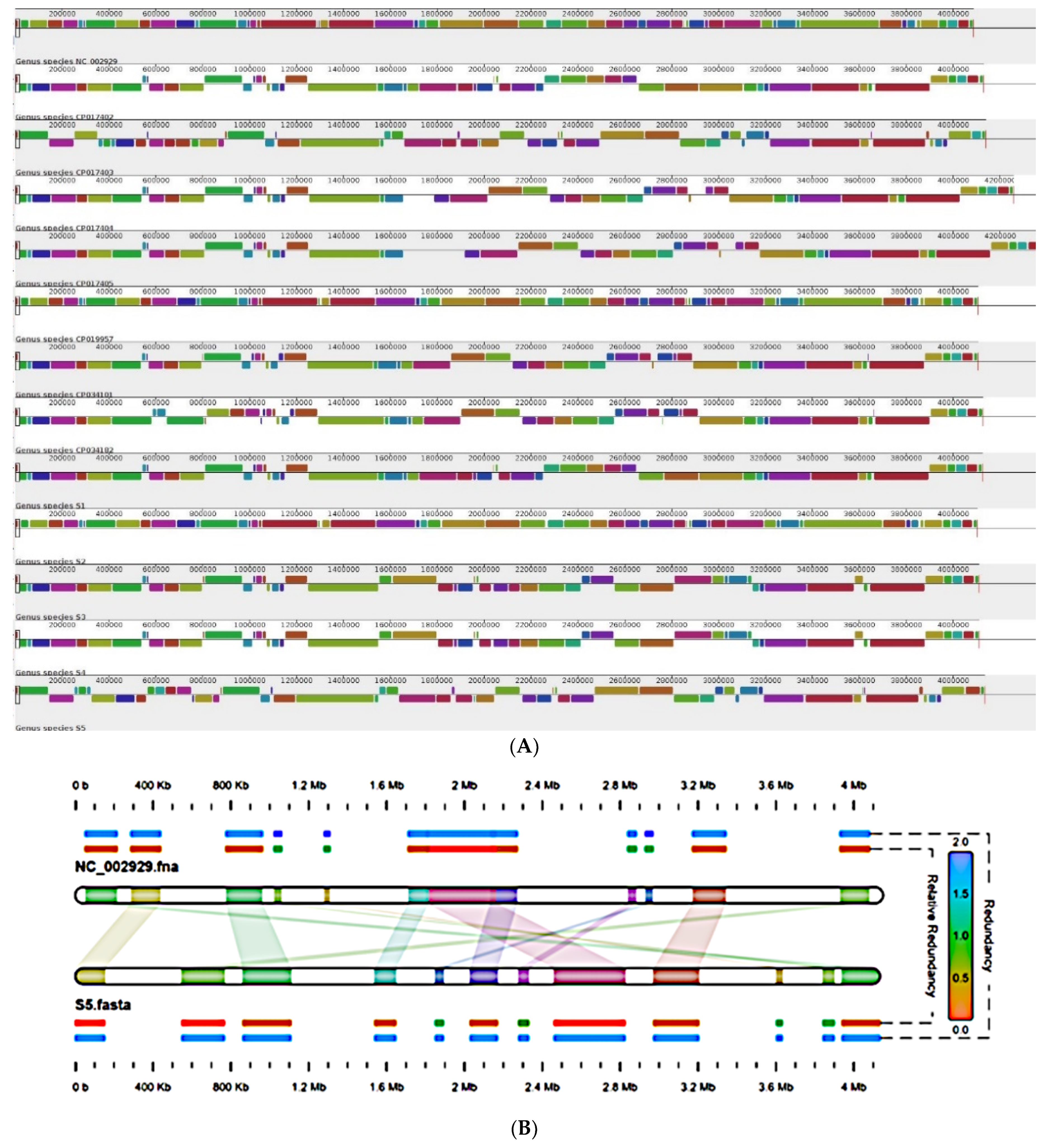
2.5. Genome Organization
2.6. Global Positioning of B. pertussis Genomes Reported from India
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. Bacterial Identification
4.3. Serotyping
4.4. DNA Extraction and Multiplex PCR
4.5. Genotyping
4.6. Whole Genome Sequencing and Annotation
4.7. Comparative Genomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gopal, D.P.; Barber, J.; Toeg, D. Pertussis (whooping cough). BMJ 2019, 364, l401. [Google Scholar] [CrossRef] [Green Version]
- Robinson, A.; Irons, L.I.; Ashworth, L.A.E. Pertussis vaccine: Present status and future prospects. Vaccine 1985, 3, 11–22. [Google Scholar] [CrossRef]
- Guiso, N.; Meade, B.D.; von König, C.H.W. Pertussis vaccines: The first hundred years. Vaccine 2019, 38, 1271–1276. [Google Scholar] [CrossRef]
- Fullen, A.R.; Yount, K.S.; Dubey, P.; Deora, R. Whoop! There it is: The surprising resurgence of pertussis. PLoS Pathog. 2020, 16, e1008625. [Google Scholar] [CrossRef]
- He, Q.; Mertsola, J. Factors contributing to pertussis resurgence. Future Microbiol. 2008, 3, 329–339. [Google Scholar] [CrossRef]
- Higgs, R.; Higgins, S.C.; Ross, P.J.; Mills, K.H.G. Immunity to the respiratory pathogen Bordetella pertussis. Mucosal Immunol. 2012, 5, 485–500. [Google Scholar] [CrossRef] [Green Version]
- Ausiello, C.M.; Cassone, A. Acellular Pertussis Vaccines and Pertussis Resurgence: Revise or Replace? mBio 2014, 5, e01339-14. [Google Scholar] [CrossRef] [Green Version]
- Esposito, S.; Stefanelli, P.; Fry, N.K.; Fedele, G.; He, Q.; Paterson, P.; World Association of Infectious Diseases and Immu-nological Disorders (WAidid); The Vaccine Study Group of the European Society of Clinical Microbiology and Infectious Diseases (EVASG). Pertussis prevention: Reasons for resurgence, and differences in the current acellular pertussis vac-cines. Front. Immunol. 2019, 10, 1344. [Google Scholar] [CrossRef] [Green Version]
- Bouchez, V.; Hegerle, N.; Strati, F.; Njamkepo, E.; Guiso, N. New Data on Vaccine Antigen Deficient Bordetella pertussis Isolates. Vaccines 2015, 3, 751–770. [Google Scholar] [CrossRef]
- Barkoff, A.-M.; Mertsola, J.; Piérard, D.; Dalby, T.; Hoegh, S.V.; Guillot, S.; Stefanelli, P.; Van Gent, M.; Berbers, G.; Vestrheim, D.; et al. Pertactin-deficient Bordetella pertussis isolates: Evidence of increased circulation in Europe, 1998 to 2015. Eurosurveillance 2019, 24, 1700832. Available online: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2019.24.7.1700832 (accessed on 11 June 2022). [CrossRef]
- Mooi, F.R.; Van Der Maas, N.A.T.; De MELKER, H.E. Pertussis resurgence: Waning immunity and pathogen adaptation—Two sides of the same coin. Epidemiol. Infect. 2014, 142, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Bart, M.J.; Harris, S.R.; Advani, A.; Arakawa, Y.; Bottero, D.; Bouchez, V.; Cassiday, P.K.; Chiang, C.-S.; Dalby, T.; Fry, N.; et al. Global Population Structure and Evolution of Bordetella pertussis and Their Relationship with Vaccination. MBio 2014, 5, e01074-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packard, E.R.; Parton, R.; Coote, J.G.; Fry, N. Sequence variation and conservation in virulence-related genes of Bordetella pertussis isolates from the UK. J. Med. Microbiol. 2004, 53, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Borisova, O.; Kombarova, S.Y.; Zakharova, N.S.; van Gent, M.; Aleshkin, V.A.; Mazurova, I.; Mooi, F.R. Antigenic Divergence between Bordetella pertussis Clinical Isolates from Moscow, Russia, and Vaccine Strains. Clin. Vaccine Immunol. 2007, 14, 234–238. [Google Scholar] [CrossRef] [Green Version]
- Sealey, K.L.; Harris, S.R.; Fry, N.K.; Hurst, L.D.; Gorringe, A.R.; Parkhill, J.; Preston, A. Genomic Analysis of Isolates From the United Kingdom 2012 Pertussis Outbreak Reveals That Vaccine Antigen Genes Are Unusually Fast Evolving. J. Infect. Dis. 2015, 212, 294–301. [Google Scholar] [CrossRef] [Green Version]
- Lahariya, C. A brief history of vaccines & vaccination in India. Indian J. Med. Res. 2014, 139, 491–511. [Google Scholar]
- Alai, S.; Ghattargi, V.; Gautam, M.; Dhotre, D.P.; Patel, K.; Pawar, S.P.; Kumar, R.; Shaligram, U.; Deobagkar, D.; Gairola, S. Genome Sequence of Bordetella pertussis Vaccine Strain BP 165. Microbiol. Resour. Announc. 2019, 8, e00150-19. [Google Scholar] [CrossRef] [Green Version]
- Alai, S.; Ghattargi, V.; Gautam, M.; Patel, K.; Pawar, S.P.; Dhotre, D.P.; Shaligram, U.; Gairola, S. Comparative genomics of whole-cell pertussis vaccine strains from India. BMC Genom. 2020, 21, 345. [Google Scholar] [CrossRef]
- Mooi, F.R.; Hallander, H.; Von König, C.W.; Hoet, B.; Guiso, N. Epidemiological Typing of Bordetella pertussis Isolates: Recommendations for a Standard Methodology. Eur. J. Clin. Microbiol. 2000, 19, 174–181. [Google Scholar] [CrossRef]
- Robinson, A.; Ashworth, L.; Irons, L. Serotyping Bordetella pertussis strains. Vaccine 1989, 7, 491–494. [Google Scholar] [CrossRef]
- Stanbridge, T.N.; Preston, N.W. Variation of serotype in strains of Bordetella pertussis. J. Hyg. 1974, 73, 305–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, A.; Parkhill, J.; Maskell, D.J. The Bordetellae: Lessons from genomics. Nat. Rev. Genet. 2004, 2, 379–390. [Google Scholar] [CrossRef]
- Bahmanjeh, A.; Noofeli, M.; Khaki, P.; Hassanzadeh, S.M. Genetic analysis of clinical and vaccine strains of Bordetella pertussis by Pulsed-Field Gel Elec-trophoresis (PFGE), Multi Locus Sequence Typing (MLST) and serotyping. Comp. Immunol. Microbiol. Fectious Dis. 2019, 64, 168–175. [Google Scholar] [CrossRef]
- Ring, N.; Abrahams, J.S.; Jain, M.; Olsen, H.; Preston, A.; Bagby, S. Resolving the complex Bordetella pertussis genome using barcoded nanopore sequencing. Microb. Genom. 2018, 4, e000234. Available online: https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000234 (accessed on 23 February 2022). [CrossRef] [PubMed]
- Bouchez, V.; Baines, S.; Guillot, S.; Brisse, S. Complete Genome Sequences of Bordetella pertussis Clinical Isolate FR5810 and Reference Strain Tohama from Combined Oxford Nanopore and Illumina Sequencing. Microbiol. Resour. Announc. 2018, 7, e01207-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Zee, A.; Schellekens, J.F.P.; Mooi, F.R. Laboratory Diagnosis of Pertussis. Clin. Microbiol. Rev. 2015, 28, 1005–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulbert, R.R.; Cotter, P.A. Laboratory Maintenance ofBordetella pertussis. Curr. Protoc. Microbiol. 2009, 15, 4B.1.1–4B.1.9. [Google Scholar] [CrossRef]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.; Lynch, S.V. Use of 16S rRNA Gene for Identification of a Broad Range of Clinically Relevant Bacterial Pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef]
- Zintgraff, J.; Irazu, L.; Lara, C.S.; Rodriguez, M.; Santos, M. The classical Bordetella species and MALDI-TOF technology: A brief experience. J. Med. Microbiol. 2018, 67, 1737–1742. [Google Scholar] [CrossRef]
- Leite, D.; Cassiday, P.K.; Tatti, K.M.; Vaz, T.M.I.; Tondella, M.L. Serotypes and genetic profiles of Bordetella pertussis strains isolated in the city of São Paulo, 2006–2008. J. Pediatr. 2012, 88, 357–360. Available online: http://www.jped.com.br/Redirect.aspx?varArtigo=2313 (accessed on 23 February 2022). [CrossRef] [Green Version]
- Mooi, F.R.; van Loo, I.H.; van Gent, M.; He, Q.; Bart, M.J.; Heuvelman, K.J.; de Greeff, S.C.; Diavatopoulos, D.; Teunis, P.; Nagelkerke, N.; et al. Bordetella pertussis Strains with Increased Toxin Production Associated with Pertussis Resurgence. Emerg. Infect. Dis. 2009, 15, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Etskovitz, H.; Anastasio, N.; Green, E.; May, M. Role of Evolutionary Selection Acting on Vaccine Antigens in the Re-Emergence of Bordetella Pertussis. Diseases 2019, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dienstbier, A.; Amman, F.; Petráčková, D.; Štipl, D.; Čapek, J.; Zavadilová, J.; Fabiánová, K.; Držmíšek, J.; Kumar, D.; Wildung, M.; et al. Comparative Omics Analysis of Historic and Recent Isolates of Bordetella pertussis and Effects of Genome Rearrangements on Evolution. Emerg. Infect. Dis. 2021, 27, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Weigand, M.R.; Peng, Y.; Batra, D.; Burroughs, M.; Davis, J.K.; Knipe, K.; Loparev, V.N.; Johnson, T.; Juieng, P.; Rowe, L.A.; et al. Conserved Patterns of Symmetric Inversion in the Genome Evolution of Bordetella Respiratory Pathogens. Msystems 2019, 4, e00702-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efendi, Y.S.; Susanti, D.; Tritama, E.; Pasier, M.L.; Putri, G.N.N.; Raharso, S.; Iskandar; Aditiawati, P.; Giri-Rachman, E.A.; Mukhopadhyay, B.; et al. Complete Genome Sequence of Bordetella pertussis Pelita III, the Production Strain for an Indonesian Whole-Cell Pertussis Vaccine. Genome Announc. 2017, 5, e00235-17. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.C.; Anandan, S.; Ragupathi, N.K.D.; Sethuvel, D.P.M.; Vasudevan, K.; Kumar, D.; Gupta, S.K.; Sangal, L.; Veeraraghavan, B. Genetic Diversity of Clinical Bor-detella Pertussis ST2 Strains in comparison with Vaccine Reference Strains of India. J. Genom. 2021, 9, 38–42. [Google Scholar] [CrossRef]
- Jung, S.-O.; Moon, Y.M.; Kim, S.-H.; Sung, H.Y.; Kwon, S.-J.; Kang, Y.H.; Yu, J.Y. Multilocus Sequence Analysis of Housekeeping Genes and Antigenic Determinant Genes in Bordetella pertussis Strains Isolated in Korea. Osong Public Health Res. Perspect. 2011, 2, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Kilgore, P.E.; Salim, A.M.; Zervos, M.J.; Schmitt, H.-J. Pertussis: Microbiology, Disease, Treatment, and Prevention. Clin. Microbiol. Rev. 2016, 29, 449–486. [Google Scholar] [CrossRef] [Green Version]
- WHO I Vaccines and Biologicals. Laboratory Manual for the Diagnosis of Whooping Cough Caused by Bordetella pertussis/Bordetella parapertussis. 2014. Available online: www.who.int/vaccines-documents/ (accessed on 23 February 2022).
- Parkhill, J.; Sebaihia, M.; Preston, A.; Murphy, L.; Thomson, N.; Harris, D.E.; Holden, M.; Churcher, C.M.; Bentley, S.D.; Mungall, K.L.; et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003, 35, 32–40. [Google Scholar] [CrossRef]
- King, A.J.; van der Lee, S.; Mohangoo, A.; van Gent, M.; van der Ark, A.; van de Waterbeemd, B. Genome-Wide Gene Expression Analysis of Bordetella pertussis Isolates Associated with a Resurgence in Pertussis: Elucidation of Factors Involved in the Increased Fitness of Epidemic Strains. PLoS ONE 2013, 8, e66150. [Google Scholar] [CrossRef] [Green Version]
- Lam, C.; Octavia, S.; Bahrame, Z.; Sintchenko, V.; Gilbert, G.L.; Lan, R. Selection and emergence of pertussis toxin promoter ptxP3 allele in the evolution of Bordetella pertussis. Infect. Genet. Evol. 2012, 12, 492–495. [Google Scholar] [CrossRef] [PubMed]
- Preston, A. The role of B. pertussis vaccine antigen gene variants in pertussis resurgence and possible consequences for vaccine development. Hum. Vaccines Immunother. 2016, 12, 1274–1276. [Google Scholar] [CrossRef] [Green Version]
- Gzyl, A.; Augustynowicz, E.; van Loo, I.; Ślusarczyk, J. Temporal nucleotide changes in pertactin and pertussis toxin genes in Bordetella pertussis strains isolated from clinical cases in Poland. Vaccine 2001, 20, 299–303. [Google Scholar] [CrossRef]
- Mosiej, E.; Augustynowicz, E.; Zawadka, M.; Dąbrowski, W.; Lutyńska, A. Strain Variation among Bordetella pertussis Isolates Circulating in Poland after 50 Years of Whole-Cell Pertussis Vaccine Use. J. Clin. Microbiol. 2011, 49, 1452–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez-Ferman, J.L.; Villarreal-Treviño, L.; Ramírez-Aranda, J.M.; Camacho-Ortiz, A.; Ballesteros-Elizondo, M.R.; Moreno-Juárez, M.R.; Mendoza-Olazarán, S.; de la OCavazos, M.E.; Villarreal-Pérez, J.Z.; Gómez-Govea, M.A.; et al. Emerging of ptxP 3 lineage in B ordetella pertussis strains circulating in a population in northeastern Mexico. Epidemiol. Infect. 2018, 146, 2096–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriuchi, T.; Vichit, O.; Vutthikol, Y.; Hossain, S.; Samnang, C.; Toda, K.; Grabovac, V.; Hiramatsu, Y.; Otsuka, N.; Shibayama, K.; et al. Molecular epidemiology of Bordetella pertussis in Cambodia determined by direct genotyping of clinical specimens. Int. J. Infect. Dis. 2017, 62, 56–58. [Google Scholar] [CrossRef] [Green Version]
- Andrade, B.; Marín, M.A.; Cambuy, D.D.; Fonseca, E.L.; Souza, N.F.; Vicente, A.C.P. Complete genome sequence of a clinical Bordetella pertussis isolate from Brazil. Memórias Inst. Oswaldo Cruz 2014, 109, 972–974. [Google Scholar] [CrossRef] [Green Version]
- Rocha, E.L.; Leite, D.; Camargo, C.H.; Martins, L.M.; Silva, R.S.N.; Martins, V.P.; Campos, T.A. The characterization of Bordetella pertussis strains isolated in the Central-Western region of Brazil suggests the selection of a specific genetic profile during 2012–2014 outbreaks. Epidemiol. Infect. 2017, 145, 1392–1397. [Google Scholar] [CrossRef] [Green Version]
- Safarchi, A.; Octavia, S.; Nikbin, V.S.; Lotfi, M.N.; Zahraei, S.M.; Tay, C.Y.; Lamichhane, B.; Shahcheraghi, F.; Lan, R. Genomic epidemiology of Iranian Bordetella pertussis: 50 years after the implementation of whole cell vaccine. Emerg. Microbes Infect. 2019, 8, 1416–1427. [Google Scholar] [CrossRef] [Green Version]
- Haghighi, F.; Shahcheraghi, F.; Abbasi, E.; Eshraghi, S.S.; Zeraati, H.; Mousavi, S.A.J.; Asgarian-Omran, H.; Douraghi, M.; Shokri, F. Genetic Profile Variation in Vaccine Strains and Clinical Isolates of Bordetella pertussis Recovered from Iranian Patients. Avicenna J. Med. Biotechnol. 2014, 6, 178–184. [Google Scholar]
- Vashishtha, V.M.; Bansal, C.P.; Gupta, S.G. Pertussis vaccines: Position paper of Indian Academy of Pediatrics (IAP). Indian Pediatr. 2013, 50, 1001–1009. [Google Scholar] [CrossRef]
- Bar-On, E.S.; Goldberg, E.; Hellmann, S.; Leibovici, L. Combined DTP-HBV-HIB vaccine versus separately administered DTP-HBV and HIB vaccines for primary prevention of diphtheria, tetanus, pertussis, hepatitis B and Haemophilus influenzae B (HIB). Cochrane Database Syst. Rev. 2012, 4, CD005530. [Google Scholar] [CrossRef]
- Dorji, D.; Mooi, F.; Yantorno, O.; Deora, R.; Graham, R.M.; Mukkur, T.K. Bordetella Pertussis virulence factors in the continuing evolution of whooping cough vaccines for improved performance. Med. Microbiol. Immunol. 2017, 207, 3–26. [Google Scholar] [CrossRef]
- Breakwell, L.; Kelso, P.; Finley, C.; Schoenfeld, S.; Goode, B.; Misegades, L.K.; Martin, S.W.; Acosta, A.M. Pertussis Vaccine Effectiveness in the Setting of Pertactin-Deficient Pertussis. Pediatrics 2016, 137, e20153973. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Luan, Y.; Du, Q.; Shu, C.; Peng, X.; Wei, H.; Hou, T.; Liu, Y.; Liu, X.; Li, Y. The global prevalence ptxP3 lineage of Bordetella pertussis was rare in young children with the co-purified aPV vaccination: A 5 years retrospective study. BMC Infect. Dis. 2020, 20, 615. [Google Scholar] [CrossRef]
- Mooi, F.R.; Zeddeman, A.; van Gent, M. The pertussis problem: Classical epidemiology and strain characterization should go hand in hand. J. Pediatr. 2015, 91, 315–317. [Google Scholar] [CrossRef] [Green Version]
- van Gent, M.; Bart, M.J.; van der Heide, H.G.J.; Heuvelman, K.J.; Mooi, F.R. Small Mutations in Bordetella pertussis Are Associated with Selective Sweeps. PLoS ONE 2012, 7, e46407. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, S.; Gautam, M.; Gairola, S. Role of vaccine manufacturers in developing countries towards global healthcare by providing quality vaccines at affordable prices. Clin. Microbiol. Infect. 2014, 20, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.; Pereira, A.S.P.; Moreira-Filho, C.A.; Bando, S.Y.; Tambourgi, D.V. Comparative analysis of a Bordetella pertussis patient isolated strain and classical strains used in the pertussis vaccine. Vaccine 2005, 23, 4353–4358. [Google Scholar] [CrossRef]
- Queenan, A.M.; Fernandez, J.; Shang, W.; Wiertsema, S.; Dobbelsteen, G.P.J.M.V.D.; Poolman, J. The mouse intranasal challenge model for potency testing of whole-cell pertussis vaccines. Expert Rev. Vaccines 2014, 13, 1265–1270. [Google Scholar] [CrossRef]
- Merkel, T.J.; Halperin, S.A. Nonhuman Primate and Human Challenge Models of Pertussis. J. Infect. Dis. 2014, 209, S20–S23. [Google Scholar] [CrossRef] [Green Version]
- Xing, D.; Das, R.; O’Neill, T.; Corbel, M.; Dellepiane, N.; Milstien, J. Laboratory testing of whole cell pertussis vaccine: A WHO proficiency study using the Kendrick test. Vaccine 2001, 20, 342–351. [Google Scholar] [CrossRef]
- De Graaf, H.; Ibrahim, M.; Hill, A.; Gbesemete, D.; Gorringe, A.; Diavatopoulos, D.; Kester, K.; Berbers, G.; Faust, S.; Read, R. 167. A Bordetella pertussis Human Challenge Model Induces Immunizing Colonization in the Absence of Symptoms. Open Forum Infect. Dis. 2018, 5 (Suppl. S1), S17. [Google Scholar] [CrossRef]
- Ebell, M.H.; Marchello, C.; Callahan, M. Clinical Diagnosis of Bordetella Pertussis Infection: A Systematic Review. J. Am. Board Fam. Med. 2017, 30, 308–319. [Google Scholar] [CrossRef] [Green Version]
- Kurzynski, T.A.; Boehm, D.M.; Rott-Petri, J.A.; Schell, R.F.; Allison, P.E. Comparison of modified Bordet-Gengou and modified Re-gan-Lowe media for the isolation of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 1988, 26, 2661–2663. [Google Scholar] [CrossRef] [Green Version]
- Advani, A.; Donnelly, D.; Hallander, H. Reference System for Characterization of Bordetella pertussis Pulsed-Field Gel Electro-phoresis Profiles. J. Clin. Microbiol. 2004, 42, 2890–2897. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rear-rangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
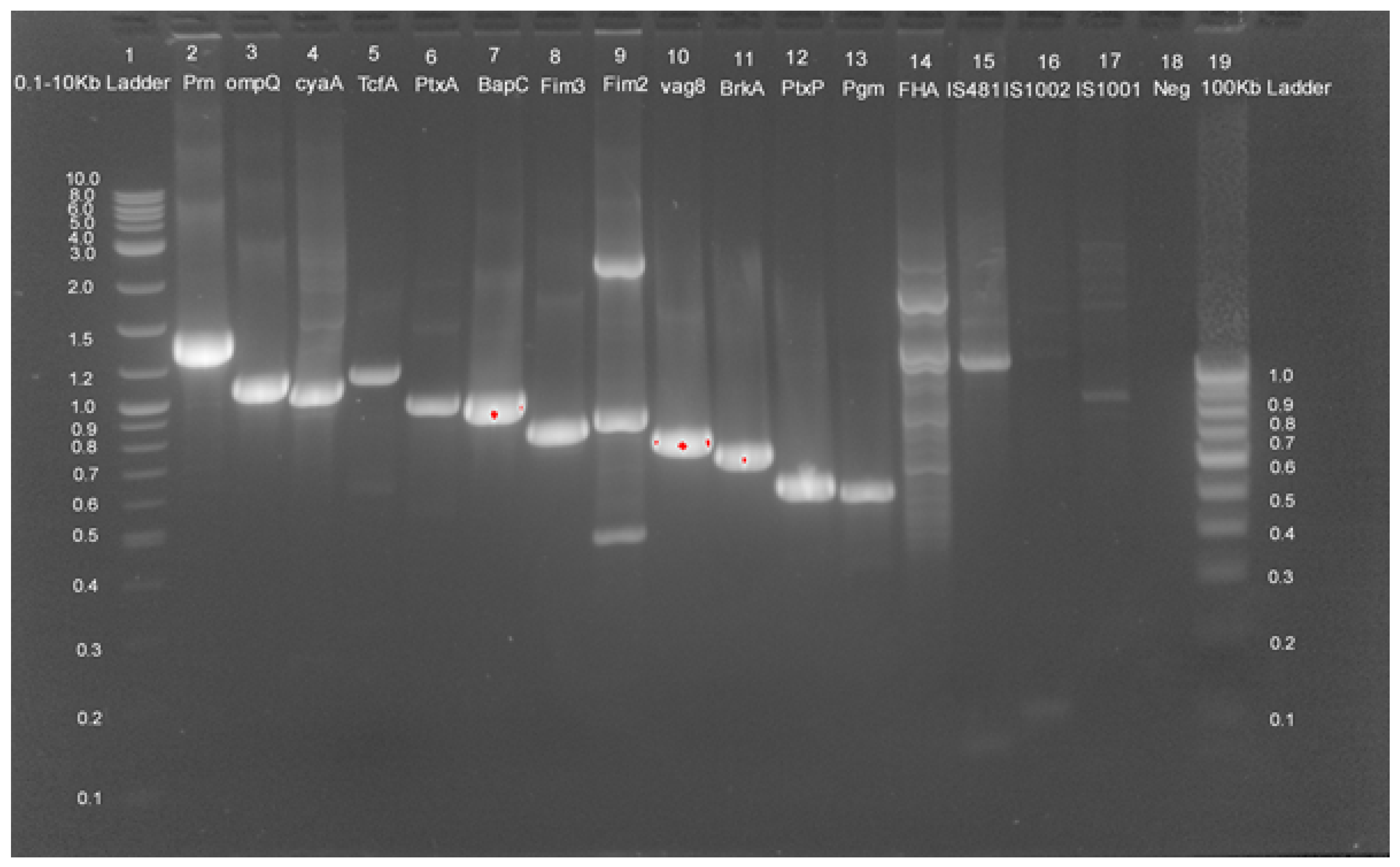

| Sample | Place | Year | Age | Gender | Serotyping (Fim2/Fim3) | Genome Size | Accession No |
|---|---|---|---|---|---|---|---|
| S1 | Pune | 2018 | 3.5 | Female | +/+ | 4124562 | CP077657 |
| S2 | Pune | 2019 | 3 | Male | +/+ | 4102989 | CP077656 |
| S3 | Pune | 2019 | 13 | Female | +/+ | 4109561 | CP077655 |
| S4 | Pune | 2020 | 02 | Female | +/+ | 4109559 | CP077654 |
| S5 | Pune | 2020 | 1 | Male | +/+ | 4135302 | CP077653 |
| Strain | Accession Number | Feature | ST | PtxP | PtxA | Prn | Fim3 | Fim2 |
|---|---|---|---|---|---|---|---|---|
| TohamaI | NC2099 | Reference Strain | 2 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| J445 | CP017402 | Vaccine Strain | 2 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| J446 | CP017403 | Vaccine Strain | 1 | PtxP2 | PtxA4 | Prn7 | Fim 3-1 | Fim 2-2 |
| J447 | CP017404 | Vaccine Strain | 2 | PtxP1 | PtxA1 | Prn1 | Fim 3-1 | Fim 2-1 |
| J448 | CP017405 | Vaccine Strain | 2 | PtxP1 | PtxA1 | Prn1 | Fim 3-1 | Fim 2-1 |
| Bp165 | RSFF00001 | Vaccine Strain | 2 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| Pelita III | CP019957 | Vaccine Strain | 1 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| S1 | CP077657 | Clinical Isolate | 2 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| S2 | CP077656 | Clinical Isolate | 1 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| S3 | CP077655 | Clinical Isolate | 2 | PtxP3 | PtxA2 | Prn2 | Fim 3-1 | Fim 2-1 |
| S4 | CP077654 | Clinical Isolate | 2 | PtxP3 | PtxA2 | Prn2 | Fim 3-1 | Fim 2-1 |
| S5 | CP077653 | Clinical Isolate | 2 | PtxP1 | PtxA2 | Prn1 | Fim 3-1 | Fim 2-1 |
| BPD1 | CP034182 | Clinical Isolate | 2 | PtxP1 | PtxA1 | Prn1 | Fim 3-1 | Fim 2-1 |
| BPD2 | CP034101 | Clinical Isolate | 2 | PtxP1 | PtxA1 | Prn1 | Fim3-1 | Fim2-1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alai, S.; Gautam, M.; Palkar, S.; Oswal, J.; Gairola, S.; Dhotre, D.P. Characterization of Bordetella pertussis Strains Isolated from India. Pathogens 2022, 11, 794. https://doi.org/10.3390/pathogens11070794
Alai S, Gautam M, Palkar S, Oswal J, Gairola S, Dhotre DP. Characterization of Bordetella pertussis Strains Isolated from India. Pathogens. 2022; 11(7):794. https://doi.org/10.3390/pathogens11070794
Chicago/Turabian StyleAlai, Shweta, Manish Gautam, Sonali Palkar, Jitendra Oswal, Sunil Gairola, and Dhiraj P. Dhotre. 2022. "Characterization of Bordetella pertussis Strains Isolated from India" Pathogens 11, no. 7: 794. https://doi.org/10.3390/pathogens11070794
APA StyleAlai, S., Gautam, M., Palkar, S., Oswal, J., Gairola, S., & Dhotre, D. P. (2022). Characterization of Bordetella pertussis Strains Isolated from India. Pathogens, 11(7), 794. https://doi.org/10.3390/pathogens11070794





