The Microbiome as Part of the Contemporary View of Tuberculosis Disease
Abstract
:1. Introduction
2. The Clinical Course of Tuberculosis
2.1. Definitions and Clinical Manifestations
2.2. Tuberculosis Treatment
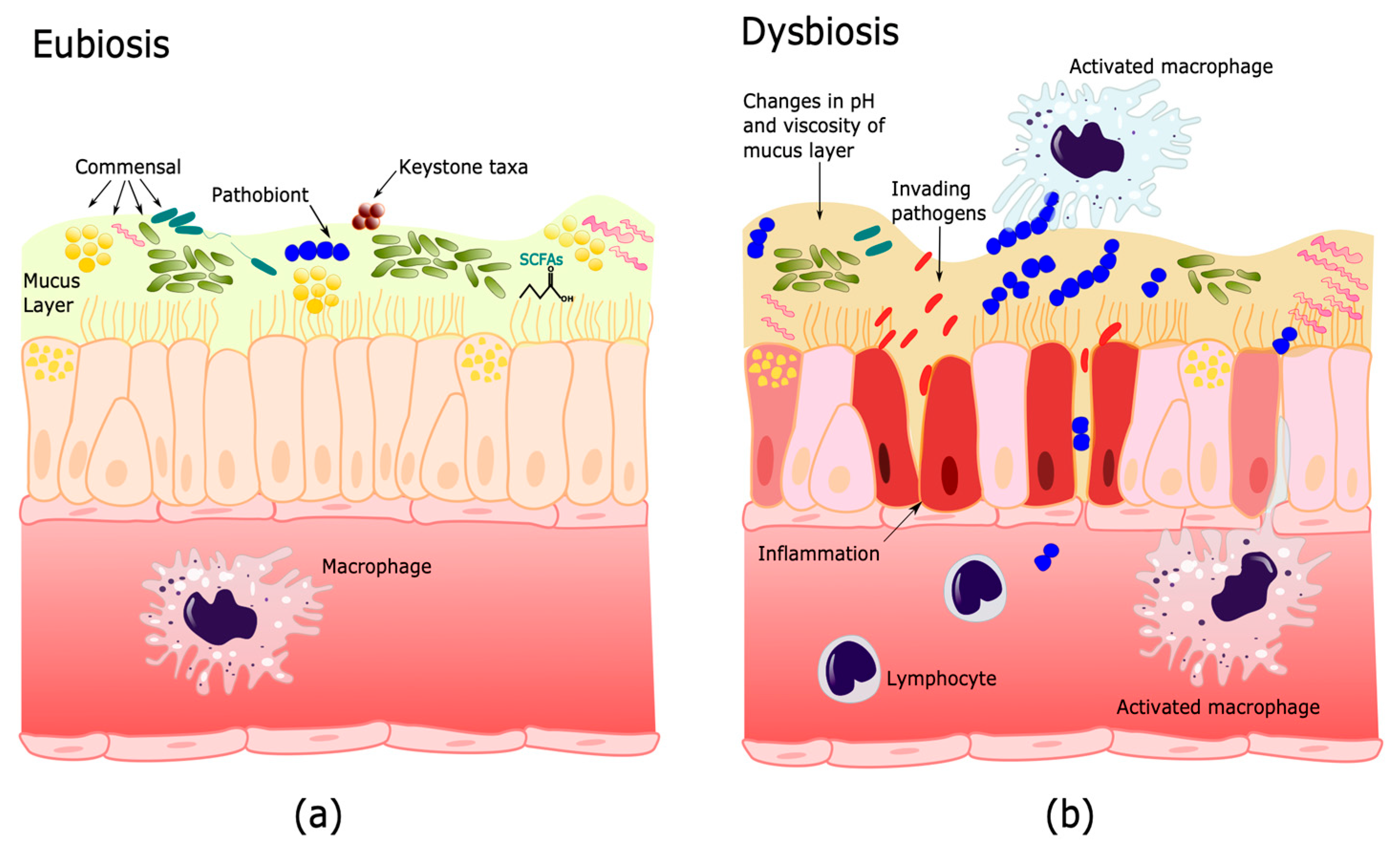
3. Human Microbiome and Its Importance in Health and Disease
3.1. Microbiome Functions
3.2. Gut–Lung Axis
3.3. The Lower Respiratory Tract (LRT) Microbiome
4. Microbiome Changes during Tuberculosis
4.1. Microbiome and Mycobacterium Tuberculosis Infection
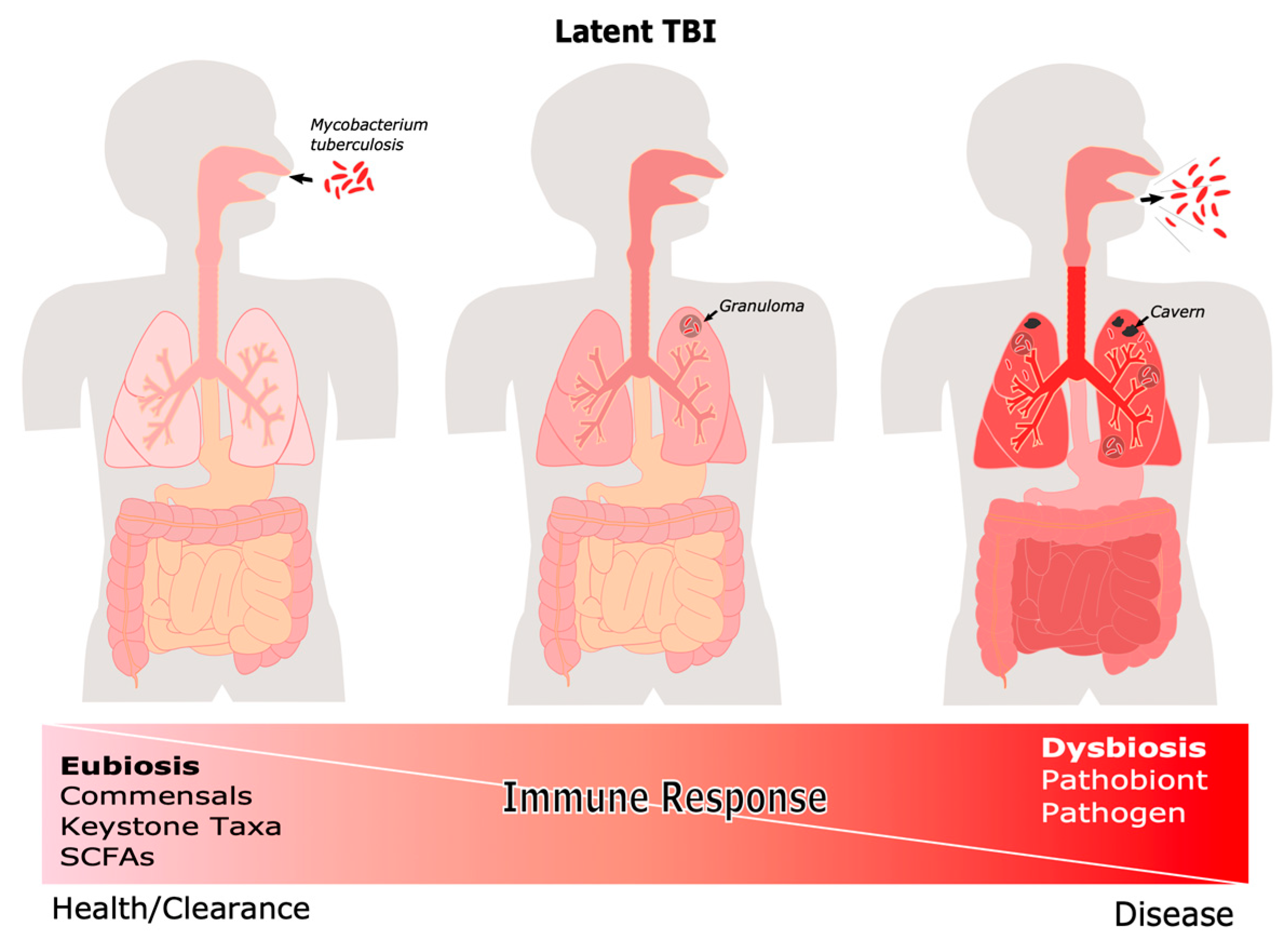
4.2. The Microbiome during Latent and Active TB
4.3. Microbiome Changes during and after Antituberculosis Treatment
4.4. Influence of the Microbiome in Post-TB Patients
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Comas, I.; Coscolla, M.; Luo, T.; Borrell, S.; Holt, K.E.; Kato-Maeda, M.; Parkhill, J.; Malla, B.; Berg, S.; Thwaites, G.; et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 2013, 45, 1176–1182. [Google Scholar] [CrossRef]
- Kay, G.L.; Sergeant, M.J.; Zhou, Z.; Chan, J.Z.M.; Millard, A.; Quick, J.; Szikossy, I.; Pap, I.; Spigelman, M.; Loman, N.J.; et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat. Commun. 2015, 6, 6717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barberis, I.; Bragazzi, N.L.; Galluzzo, L.; Martini, M. The history of tuberculosis: From the first historical records to the isolation of Koch’s bacillus. J. Prev. Med. Hyg. 2017, 58, E9. [Google Scholar] [PubMed]
- Pai, M.; Behr, M.A.; Dowdy, D.; Dheda, K.; Divangahi, M.; Boehme, C.C.; Ginsberg, A.; Swaminathan, S.; Spigelman, M.; Getahun, H.; et al. Tuberculosis. Nat. Rev. Dis. Primers 2016, 2, 16076. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2021; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Adami, A.J.; Cervantes, J.L. The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis 2015, 95, 651–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, B.-Y.; Maulén, N.P.; Adami, A.J.; Granados, H.; Balcells, M.E.; Cervantes, J. Microbiome Changes during Tuberculosis and Antituberculous Therapy. Clin. Microbiol. Rev. 2016, 29, 915–926. [Google Scholar] [CrossRef] [Green Version]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Vidyarthi, A.; Nadeem, S.; Negi, S.; Nair, G.; Agrewala, J.N. Alteration in the Gut Microbiota Provokes Susceptibility to Tuberculosis. Front. Immunol. 2016, 7, 529. [Google Scholar] [CrossRef]
- Negi, S.; Pahari, S.; Bashir, H.; Agrewala, J.N. Gut Microbiota Regulates Mincle Mediated Activation of Lung Dendritic Cells to Protect Against Mycobacterium tuberculosis. Front. Immunol. 2019, 10, 1142. [Google Scholar] [CrossRef] [Green Version]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Namasivayam, S.; Kauffman, K.D.; McCulloch, J.A.; Yuan, W.; Thovarai, V.; Mittereder, L.R.; Trinchieri, G.; Barber, D.L.; Sher, A. Correlation between Disease Severity and the Intestinal Microbiome in Mycobacterium tuberculosis-Infected Rhesus Macaques. mBio 2019, 10, e01018-19. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. Homeostasis and its disruption in the lung microbiome. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2015, 309, L1047–L1055. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.D.; Budden, K.F.; Neal, R.; Hansbro, P.M. Microbiome effects on immunity, health and disease in the lung. Clin. Transl. Immunol. 2017, 6, e133. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2016, 10, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Coburn, B.; Wang, P.W.; Diaz Caballero, J.; Clark, S.T.; Brahma, V.; Donaldson, S.; Zhang, Y.; Surendra, A.; Gong, Y.; Tullis, D.E.; et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015, 5, 10241. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [Green Version]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut–Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef] [Green Version]
- Mezouar, S.; Chantran, Y.; Michel, J.; Fabre, A.; Dubus, J.-C.; Leone, M.; Sereme, Y.; Mège, J.-L.; Ranque, S.; Desnues, B.; et al. Microbiome and the immune system: From a healthy steady-state to allergy associated disruption. Hum. Microbiome J. 2018, 10, 11–20. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.-M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef] [Green Version]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Ryndak, M.B.; Laal, S. Mycobacterium tuberculosis Primary Infection and Dissemination: A Critical Role for Alveolar Epithelial Cells. Front. Cell. Infect. Microbiol. 2019, 9, 299. [Google Scholar] [CrossRef]
- World Health Organization. WHO consolidated guidelines on tuberculosis. In Module 1: Prevention—Tuberculosis Preventive Treatment; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- World Health Organization. Systematic Screening for Active Tuberculosis: An Operational Guide; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Cadena, A.M.; Fortune, S.M.; Flynn, J.L. Heterogeneity in tuberculosis. Nat. Rev. Immunol. 2017, 17, 691–702. [Google Scholar] [CrossRef]
- Coleman, M.T.; Maiello, P.; Tomko, J.; Frye, L.J.; Fillmore, D.; Janssen, C.; Klein, E.; Lin, P.L. Early Changes by 18Fluorodeoxyglucose Positron Emission Tomography Coregistered with Computed Tomography Predict Outcome after Mycobacterium tuberculosis Infection in Cynomolgus Macaques. Infect. Immun. 2014, 82, 2400–2404. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.L.; Maiello, P.; Gideon, H.P.; Coleman, M.T.; Cadena, A.M.; Rodgers, M.A.; Gregg, R.; O’Malley, M.; Tomko, J.; Fillmore, D.; et al. PET CT Identifies Reactivation Risk in Cynomolgus Macaques with Latent M. tuberculosis. PLoS Pathog. 2016, 12, e1005739. [Google Scholar] [CrossRef]
- Newsom, S.W.B. Tuberculosis. Clinical diagnosis and management of tuberculosis and measures for its prevention and control. J. Hosp. Infect. 2006, 64, 309–310. [Google Scholar] [CrossRef]
- Rathman, G.; Sillah, J.; Hill, P.C.; Murray, J.F.; Adegbola, R.; Corrah, T.; Lienhardt, C.; McAdam, K.P. Clinical and radiological presentation of 340 adults with smear-positive tuberculosis in The Gambia. Int. J. Tuberc. Lung Dis. 2003, 7, 942–947. [Google Scholar]
- Murray, H.W.; Tuazon, C.U.; Kirmani, N.; Sheagren, J.N. The Adult Respiratory Distress Syndrome Associated with Miliary Tuberculosis. Chest 1978, 73, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Brändli, O. The clinical presentation of tuberculosis. Respiration 1998, 65, 97–105. [Google Scholar] [CrossRef]
- Loddenkemper, R.; Lipman, M.; Zumla, A. Clinical Aspects of Adult Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 6, a017848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Definitions and Reporting Framework for Tuberculosis—2013 Revision: Updated December 2014 and January 2020; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Caws, M.; Thwaites, G.; Dunstan, S.; Hawn, T.R.; Thi Ngoc Lan, N.; Thuong, N.T.T.; Stepniewska, K.; Huyen, M.N.T.; Bang, N.D.; Huu Loc, T.; et al. The Influence of Host and Bacterial Genotype on the Development of Disseminated Disease with Mycobacterium tuberculosis. PLoS Pathog. 2008, 4, e1000034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y. Diagnosis and treatment of extrapulmonary tuberculosis. Tuberc. Respir. Dis. 2015, 78, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiemersma, E.W.; van der Werf, M.J.; Borgdorff, M.W.; Williams, B.G.; Nagelkerke, N.J.D. Natural History of Tuberculosis: Duration and Fatality of Untreated Pulmonary Tuberculosis in HIV Negative Patients: A Systematic Review. PLoS ONE 2011, 6, e17601. [Google Scholar] [CrossRef]
- Nahid, P.; Dorman, S.E.; Alipanah, N.; Barry, P.M.; Brozek, J.L.; Cattamanchi, A.; Chaisson, L.H.; Chaisson, R.E.; Daley, C.L.; Grzemska, M.; et al. Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical Practice Guidelines: Treatment of Drug-Susceptible Tuberculosis. Clin. Infect. Dis. 2016, 63, e147–e195. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Treatment of Drug-Susceptible Tuberculosis and Patient Care, 2017 Update; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Dorman, S.E.; Nahid, P.; Kurbatova, E.V.; Phillips, P.P.J.; Bryant, K.; Dooley, K.E.; Engle, M.; Goldberg, S.V.; Phan, H.T.T.; Hakim, J.; et al. Four-Month Rifapentine Regimens with or without Moxifloxacin for Tuberculosis. N. Engl. J. Med. 2021, 384, 1705–1718. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Control: WHO Report 2010; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- World Health Organization. Meeting Report of the WHO Expert Consultation on the Definition of Extensively Drug-Resistant Tuberculosis, 27–29 October 2020; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- World Health Organization. WHO consolidated guidelines on tuberculosis. In Module 4: Treatment—Drug-Resistant Tuberculosis Treatment; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- World Health Organization. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Sterling, T.R.; Njie, G.; Zenner, D.; Cohn, D.L.; Reves, R.; Ahmed, A.; Menzies, D.; Horsburgh, C.R.; Crane, C.M.; Burgos, M.; et al. Guidelines for the Treatment of Latent Tuberculosis Infection: Recommendations from the National Tuberculosis Controllers Association and CDC, 2020. MMWR. Recomm. Rep. 2020, 69, 1196–1206. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Bauer, M.A.; Kainz, K.; Carmona-Gutierrez, D.; Madeo, F. Microbial wars: Competition in ecological niches and within the microbiome. Microb. Cell 2018, 5, 215–219. [Google Scholar] [CrossRef]
- Kumar, P.S. Microbial dysbiosis: The root cause of periodontal disease. J. Periodontol. 2021, 92, 1079–1087. [Google Scholar] [CrossRef]
- Tshikantwa, T.S.; Ullah, M.W.; He, F.; Yang, G. Current trends and potential applications of microbial interactions for human welfare. Front. Microbiol. 2018, 9, 1156. [Google Scholar] [CrossRef] [Green Version]
- Perry, F.; Arsenault, R.J. The study of Microbe–Host Two-way communication. Microorganisms 2022, 10, 408. [Google Scholar] [CrossRef]
- Shaiber, A.; Willis, A.D.; Delmont, T.O.; Roux, S.; Chen, L.-X.; Schmid, A.C.; Yousef, M.; Watson, A.R.; Lolans, K.; Esen, Ö.C.; et al. Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. Genome Biol. 2020, 21, 292. [Google Scholar] [CrossRef]
- Giulio, P. The intestinal microbiota: Towards a multifactorial integrative model. eubiosis and dysbiosis in morbid physical and psychological conditions. Arch. Clin. Gastroenterol. 2021, 7, 24–35. [Google Scholar] [CrossRef]
- Greenbaum, S.; Greenbaum, G.; Moran-Gilad, J.; Weintraub, A.Y. Ecological Dynamics of the vaginal microbiome in relation to health and disease. Am. J. Obstet. Gynecol. 2019, 220, 324–335. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Brüssow, H. Problems with the concept of gut microbiota dysbiosis. Microb. Biotechnol. 2019, 13, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y.; et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef]
- Gutierrez Lopez, D.E.; Lashinger, L.M.; Weinstock, G.M.; Bray, M.S. Circadian rhythms and the gut microbiome synchronize the host’s metabolic response to Diet. Cell Metab. 2021, 33, 873–887. [Google Scholar] [CrossRef]
- Das, P.; Babaei, P.; Nielsen, J. Metagenomic analysis of microbe-mediated vitamin metabolism in the human gut microbiome. BMC Genom. 2019, 20, 208. [Google Scholar] [CrossRef] [Green Version]
- Segura Munoz, R.R.; Mantz, S.; Martínez, I.; Li, F.; Schmaltz, R.J.; Pudlo, N.A.; Urs, K.; Martens, E.C.; Walter, J.; Ramer-Tait, A.E. Experimental evaluation of ecological principles to understand and modulate the outcome of bacterial strain competition in gut microbiomes. ISME J. 2022, 16, 1646–1654. [Google Scholar] [CrossRef]
- Pickard, J.M.; Núñez, G. Pathogen colonization resistance in the gut and its manipulation for improved health. Am. J. Pathol. 2019, 189, 1300–1310. [Google Scholar] [CrossRef] [Green Version]
- Duar, R.M.; Henrick, B.M.; Casaburi, G.; Frese, S.A. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: The role of B. Infantis and human milk oligosaccharides. Front. Nutr. 2020, 7, 33. [Google Scholar] [CrossRef]
- Netzker, T.; Flak, M.; Krespach, M.K.C.; Stroe, M.C.; Weber, J.; Schroeckh, V.; Brakhage, A.A. Microbial interactions trigger the production of antibiotics. Curr. Opin. Microbiol. 2018, 45, 117–123. [Google Scholar] [CrossRef]
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and their potential to modulate the microbiome and immunity. Cell. Mol. Immunol. 2020, 18, 889–904. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Stacy, A.; Andrade-Oliveira, V.; McCulloch, J.A.; Hild, B.; Oh, J.H.; Perez-Chaparro, P.J.; Sim, C.K.; Lim, A.I.; Link, V.M.; Enamorado, M.; et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627.e17. [Google Scholar] [CrossRef]
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The gut microbiome as a major regulator of the gut-skin axis. Front. Microbiol. 2018, 9, 1459. [Google Scholar] [CrossRef] [Green Version]
- Adolph, T.E.; Grander, C.; Moschen, A.R.; Tilg, H. Liver–microbiome axis in health and disease. Trends Immunol. 2018, 39, 712–723. [Google Scholar] [CrossRef]
- Bajinka, O.; Simbilyabo, L.; Tan, Y.; Jabang, J.; Saleem, S.A. Lung-Brain Axis. Crit. Rev. Microbiol. 2022, 48, 257–269. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassalle, F.; Spagnoletti, M.; Fumagalli, M.; Shaw, L.; Dyble, M.; Walker, C.; Thomas, M.G.; Bamberg Migliano, A.; Balloux, F. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to Diet. Mol. Ecol. 2017, 27, 182–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, J.R.; González-Torres, P.; Pittis, A.A.; Bejarano, L.A.; Cozzuto, L.; Andreu-Somavilla, N.; Alloza-Trabado, M.; Valentín, A.; Ksiezopolska, E.; Company, C.; et al. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome 2018, 6, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wampach, L.; Heintz-Buschart, A.; Fritz, J.V.; Ramiro-Garcia, J.; Habier, J.; Herold, M.; Narayanasamy, S.; Kaysen, A.; Hogan, A.H.; Bindl, L.; et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 2018, 9, 5091. [Google Scholar] [CrossRef] [Green Version]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung–gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef]
- Fan, Q.; Wang, H.; Mao, C.; Li, J.; Zhang, X.; Grenier, D.; Yi, L.; Wang, Y. Structure and signal regulation mechanism of interspecies and Interkingdom quorum sensing system receptors. J. Agric. Food Chem. 2022, 70, 429–445. [Google Scholar] [CrossRef]
- Mayhew, D.; Devos, N.; Lambert, C.; Brown, J.R.; Clarke, S.C.; Kim, V.L.; Magid-Slav, M.; Miller, B.E.; Ostridge, K.K.; Patel, R.; et al. Longitudinal profiling of the lung microbiome in the Aeris study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018, 73, 422–430. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, R.; Xue, Y.; Jonassen, I.; Haaland, I.; Kommedal, Ø.; Wiker, H.G.; Drengenes, C.; Bakke, P.S.; Eagan, T.M. Repeated bronchoscopy in health and obstructive lung disease: Is the airway microbiome stable? BMC Pulm. Med. 2021, 21, 342. [Google Scholar] [CrossRef]
- Marimón, J.M. The lung microbiome in health and respiratory diseases. Clin. Pulm. Med. 2018, 25, 131–137. [Google Scholar] [CrossRef]
- Wang, G.; Hu, Y.-X.; He, M.-Y.; Xie, Y.-H.; Su, W.; Long, D.; Zhao, R.; Wang, J.; Dai, C.; Li, H.; et al. Gut-lung dysbiosis accompanied by diabetes mellitus leads to pulmonary fibrotic change through the NF-ΚB signaling pathway. Am. J. Pathol. 2021, 191, 838–856. [Google Scholar] [CrossRef]
- Luo, M.; Liu, Y.; Wu, P.; Luo, D.-X.; Sun, Q.; Zheng, H.; Hu, R.; Pandol, S.J.; Li, Q.-F.; Han, Y.-P.; et al. Alternation of gut microbiota in patients with pulmonary tuberculosis. Front. Physiol. 2017, 8, 822. [Google Scholar] [CrossRef]
- Wang, Z.; Singh, R.; Miller, B.E.; Tal-Singer, R.; Van Horn, S.; Tomsho, L.; Mackay, A.; Allinson, J.P.; Webb, A.J.; Brookes, A.J.; et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 2017, 73, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Frey, D.L.; Boutin, S.; Dittrich, S.A.; Graeber, S.Y.; Stahl, M.; Wege, S.; Herth, F.J.F.; Sommerburg, O.; Schultz, C.; Mall, M.A.; et al. Relationship between airway dysbiosis, inflammation and lung function in adults with cystic fibrosis. J. Cyst. Fibros. 2021, 20, 754–760. [Google Scholar] [CrossRef]
- Dong, H.; Huang, D.; Yuan, M.; Cai, S. The characterization of lung microbiome in sputum of lung cancer patients with different clinicopathology. Am. J. Respir. Crit. Care Med. 2019, 199, A5553. [Google Scholar] [CrossRef]
- Roquilly, A.; Torres, A.; Villadangos, J.A.; Netea, M.G.; Dickson, R.; Becher, B.; Asehnoune, K. Pathophysiological role of respiratory dysbiosis in hospital-acquired pneumonia. Lancet Respir. Med. 2019, 7, 710–720. [Google Scholar] [CrossRef]
- Klomp, M.; Ghosh, S.; Mohammed, S.; Nadeem Khan, M. From virus to inflammation, how influenza promotes lung damage. J. Leukoc. Biol. 2020, 110, 115–122. [Google Scholar] [CrossRef]
- Pacheco, G.A.; Gálvez, N.M.; Soto, J.A.; Andrade, C.A.; Kalergis, A.M. Bacterial and viral coinfections with the human respiratory syncytial virus. Microorganisms 2021, 9, 1293. [Google Scholar] [CrossRef]
- Kitsios, G.D.; Yang, H.; Yang, L.; Qin, S.; Fitch, A.; Wang, X.-H.; Fair, K.; Evankovich, J.; Bain, W.; Shah, F.; et al. Respiratory tract dysbiosis is associated with worse outcomes in mechanically ventilated patients. Am. J. Respir. Crit. Care Med. 2020, 202, 1666–1677. [Google Scholar] [CrossRef]
- Minodier, L.; Charrel, R.N.; Ceccaldi, P.-E.; van der Werf, S.; Blanchon, T.; Hanslik, T.; Falchi, A. Prevalence of gastrointestinal symptoms in patients with influenza, clinical significance, and pathophysiology of human influenza viruses in faecal samples: What do we know? Virol. J. 2015, 12, 215. [Google Scholar] [CrossRef]
- Groves, H.T.; Cuthbertson, L.; James, P.; Moffatt, M.F.; Cox, M.J.; Tregoning, J.S. Respiratory disease following viral lung infection alters the murine gut microbiota. Front. Immunol. 2018, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Samuelson, D.R.; Smith, D.R.; Cunningham, K.C.; Wyatt, T.A.; Hall, S.C.; Murry, D.J.; Chhonker, Y.S.; Knoell, D.L. Zip8-mediated intestinal dysbiosis impairs pulmonary host defense against bacterial pneumonia. Int. J. Mol. Sci. 2022, 23, 1022. [Google Scholar] [CrossRef]
- Tsang, T.K.; Lee, K.H.; Foxman, B.; Balmaseda, A.; Gresh, L.; Sanchez, N.; Ojeda, S.; Lopez, R.; Yang, Y.; Kuan, G.; et al. Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin. Infect. Dis. 2019, 71, 1195–1203. [Google Scholar] [CrossRef]
- Hakim, H.; Dallas, R.; Wolf, J.; Tang, L.; Schultz-Cherry, S.; Darling, V.; Johnson, C.; Karlsson, E.A.; Chang, T.-C.; Jeha, S.; et al. Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clin. Infect. Dis. 2018, 67, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xiong, K.; Zhao, S.; Zhang, C.; Zhang, J.; Xu, L.; Ma, A. Long-term effects of multi-drug-resistant tuberculosis treatment on gut microbiota and its health consequences. Front. Microbiol. 2020, 11, 53. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Sala, C.; Benjak, A.; Goletti, D.; Banu, S.; Mazza-Stadler, J.; Jaton, K.; Busso, P.; Remm, S.; Leleu, M.; Rougemont, J.; et al. Multicenter analysis of sputum microbiota in tuberculosis patients. PLoS ONE 2020, 15, e0240250. [Google Scholar] [CrossRef] [PubMed]
- Harishankar, M.; Selvaraj, P.; Bethunaickan, R. Influence of genetic polymorphism towards pulmonary tuberculosis susceptibility. Front. Med. 2018, 5, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, A.; Corral, D.; Colom, A.; Levillain, F.; Peixoto, A.; Hudrisier, D.; Poquet, Y.; Neyrolles, O. The host microbiota contributes to early protection against lung colonization by mycobacterium tuberculosis. Front. Immunol. 2018, 9, 2656. [Google Scholar] [CrossRef]
- Namasivayam, S.; Sher, A.; Glickman, M.S.; Wipperman, M.F. The microbiome and tuberculosis: Early evidence for cross talk. mBio 2018, 9, e01420-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, P.; Jain, A.; Bisen, P.S. Microbiome diversity in the sputum of patients with pulmonary tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1205–1210. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.-y.; Paulson, J.N.; Stine, O.C.; Weinstock, G.M.; Cervantes, J.L. Meta-analysis of the lung microbiota in pulmonary tuberculosis. Tuberculosis 2018, 109, 102–108. [Google Scholar] [CrossRef]
- Vázquez-Pérez, J.A.; Carrillo, C.O.; Iñiguez-García, M.A.; Romero-Espinoza, I.; Márquez-García, J.E.; Falcón, L.I.; Torres, M.; Herrera, M.T. Alveolar microbiota profile in patients with human pulmonary tuberculosis and interstitial pneumonia. Microb. Pathog. 2020, 139, 103851. [Google Scholar] [CrossRef]
- Valdez-Palomares, F.; Muñoz Torrico, M.; Palacios-González, B.; Soberón, X.; Silva-Herzog, E. Altered microbial composition of drug-sensitive and drug-resistant TB patients compared with healthy volunteers. Microorganisms 2021, 9, 1762. [Google Scholar] [CrossRef]
- Winglee, K.; Eloe-Fadrosh, E.; Gupta, S.; Guo, H.; Fraser, C.; Bishai, W. Aerosol mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS ONE 2014, 9, e97048. [Google Scholar] [CrossRef]
- Wipperman, M.F.; Fitzgerald, D.W.; Juste, M.A.; Taur, Y.; Namasivayam, S.; Sher, A.; Bean, J.M.; Bucci, V.; Glickman, M.S. Antibiotic treatment for tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci. Rep. 2017, 7, 10767. [Google Scholar] [CrossRef]
- Balcells, M.E.; Yokobori, N.; Hong, B.-y.; Corbett, J.; Cervantes, J. The lung microbiome, vitamin D, and the tuberculous granuloma: A balance triangle. Microb. Pathog. 2019, 131, 158–163. [Google Scholar] [CrossRef]
- Perry, S.; de Jong, B.C.; Solnick, J.V.; de la Luz Sanchez, M.; Yang, S.; Lin, P.L.; Hansen, L.M.; Talat, N.; Hill, P.C.; Hussain, R.; et al. Infection with helicobacter pylori is associated with protection against tuberculosis. PLoS ONE 2010, 5, e8804. [Google Scholar] [CrossRef] [Green Version]
- Majlessi, L.; Sayes, F.; Bureau, J.-F.; Pawlik, A.; Michel, V.; Jouvion, G.; Huerre, M.; Severgnini, M.; Consolandi, C.; Peano, C.; et al. Colonization with helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of mycobacterium tuberculosis. Mucosal Immunol. 2017, 10, 1178–1189. [Google Scholar] [CrossRef]
- Namasivayam, S.; Diarra, B.; Diabate, S.; Sarro, Y.D.S.; Kone, A.; Kone, B.; Tolofoudie, M.; Baya, B.; Diakite, M.T.; Kodio, O.; et al. Patients infected with mycobacterium africanum versus mycobacterium tuberculosis possess distinct intestinal microbiota. PLoS Negl. Trop. Dis. 2020, 14, e0008230. [Google Scholar] [CrossRef]
- Cui, Z.; Zhou, Y.; Li, H.; Zhang, Y.; Zhang, S.; Tang, S.; Guo, X. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol. 2012, 12, 276. [Google Scholar] [CrossRef] [Green Version]
- Maji, A.; Misra, R.; Dhakan, D.B.; Gupta, V.; Mahato, N.K.; Saxena, R.; Mittal, P.; Thukral, N.; Sharma, E.; Singh, A.; et al. Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ. Microbiol. 2017, 20, 402–419. [Google Scholar] [CrossRef]
- Lin, D.; Wang, X.; Li, Y.; Wang, W.; Li, Y.; Yu, X.; Lin, B.; Chen, Y.; Lei, C.; Zhang, X.; et al. Sputum microbiota as a potential diagnostic marker for multidrug-resistant tuberculosis. Int. J. Med. Sci. 2021, 18, 1935–1945. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Shi, G.; Han, W.; Zhao, H.; Zhang, H.; Xi, X. Determinants of multidrug-resistant tuberculosis in Henan province in China: A case control study. BMC Public Health 2015, 16, 42. [Google Scholar] [CrossRef] [Green Version]
- Cervantes, J.; Hong, B.-Y. The gut–lung axis in tuberculosis. Pathog. Dis. 2017, 75, ftx097. [Google Scholar] [CrossRef] [Green Version]
- Namasivayam, S.; Maiga, M.; Yuan, W.; Thovarai, V.; Costa, D.L.; Mittereder, L.R.; Wipperman, M.F.; Glickman, M.S.; Dzutsev, A.; Trinchieri, G.; et al. Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome 2017, 5, 71. [Google Scholar] [CrossRef]
- Ma, X.; Shin, Y.-J.; Jang, H.-M.; Joo, M.-K.; Yoo, J.-W.; Kim, D.-H. Lactobacillus rhamnosus and Bifidobacterium Longum alleviate colitis and cognitive impairment in mice by regulating IFN-γ to IL-10 and TNF-α to IL-10 expression ratios. Sci. Rep. 2021, 11, 20659. [Google Scholar] [CrossRef]
- Lev-Sagie, A.; Goldman-Wohl, D.; Cohen, Y.; Dori-Bachash, M.; Leshem, A.; Mor, U.; Strahilevitz, J.; Moses, A.E.; Shapiro, H.; Yagel, S.; et al. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat. Med. 2019, 25, 1500–1504. [Google Scholar] [CrossRef]
- Hsu, D.; Irfan, M.; Jabeen, K.; Iqbal, N.; Hasan, R.; Migliori, G.B.; Zumla, A.; Visca, D.; Centis, R.; Tiberi, S. Post tuberculosis treatment infectious complications. Int. J. Infect. Dis. 2020, 92, S41–S45. [Google Scholar] [CrossRef] [Green Version]
- Romanowski, K.; Balshaw, R.F.; Benedetti, A.; Campbell, J.R.; Menzies, D.; Ahmad Khan, F.; Johnston, J.C. Predicting tuberculosis relapse in patients treated with the standard 6-month regimen: An individual patient data meta-analysis. Thorax 2018, 74, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, W.; He, L.; Huang, F.; Chen, J.; Cui, P.; Shen, Y.; Zhao, J.; Wang, W.; Zhang, Y.; et al. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS ONE 2013, 8, e83445. [Google Scholar] [CrossRef]
- Naidoo, C.C.; Nyawo, G.R.; Wu, B.G.; Walzl, G.; Warren, R.M.; Segal, L.N.; Theron, G. The microbiome and tuberculosis: State of the art, potential applications, and defining the Clinical Research Agenda. Lancet Respir. Med. 2019, 7, 892–906. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa-Amezcua, M.; Galeana-Cadena, D.; Alvarado-Peña, N.; Silva-Herzog, E. The Microbiome as Part of the Contemporary View of Tuberculosis Disease. Pathogens 2022, 11, 584. https://doi.org/10.3390/pathogens11050584
Barbosa-Amezcua M, Galeana-Cadena D, Alvarado-Peña N, Silva-Herzog E. The Microbiome as Part of the Contemporary View of Tuberculosis Disease. Pathogens. 2022; 11(5):584. https://doi.org/10.3390/pathogens11050584
Chicago/Turabian StyleBarbosa-Amezcua, Martín, David Galeana-Cadena, Néstor Alvarado-Peña, and Eugenia Silva-Herzog. 2022. "The Microbiome as Part of the Contemporary View of Tuberculosis Disease" Pathogens 11, no. 5: 584. https://doi.org/10.3390/pathogens11050584
APA StyleBarbosa-Amezcua, M., Galeana-Cadena, D., Alvarado-Peña, N., & Silva-Herzog, E. (2022). The Microbiome as Part of the Contemporary View of Tuberculosis Disease. Pathogens, 11(5), 584. https://doi.org/10.3390/pathogens11050584





