Peripheral Blood Markers Correlate with the Progression of Active Tuberculosis Relative to Latent Control of Mycobacterium tuberculosis Infection in Macaques
Abstract
1. Introduction
2. Results
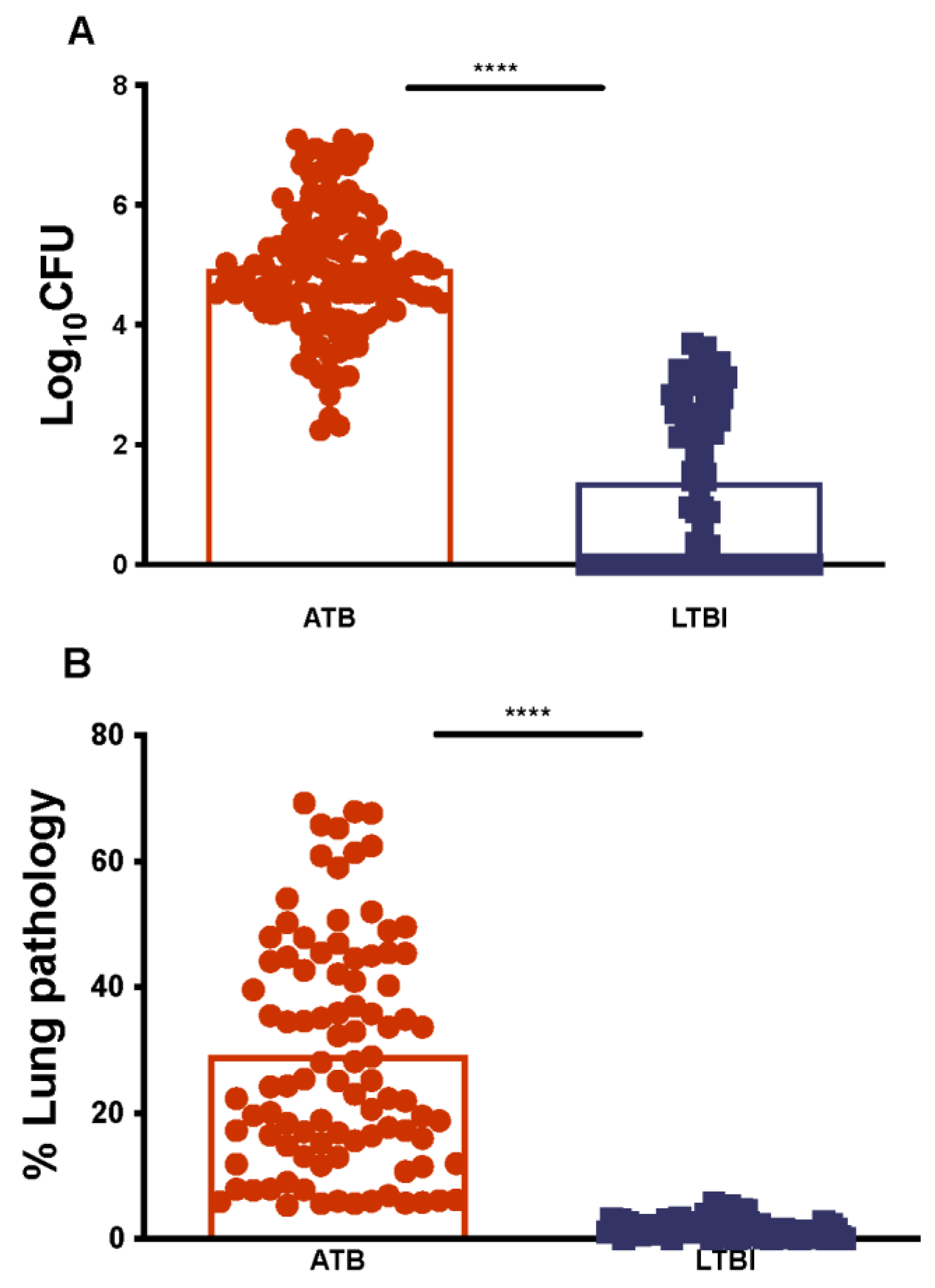
2.1. Establishing Lung Mtb Burdens and Pathology for ATB and LTBI in NHPs
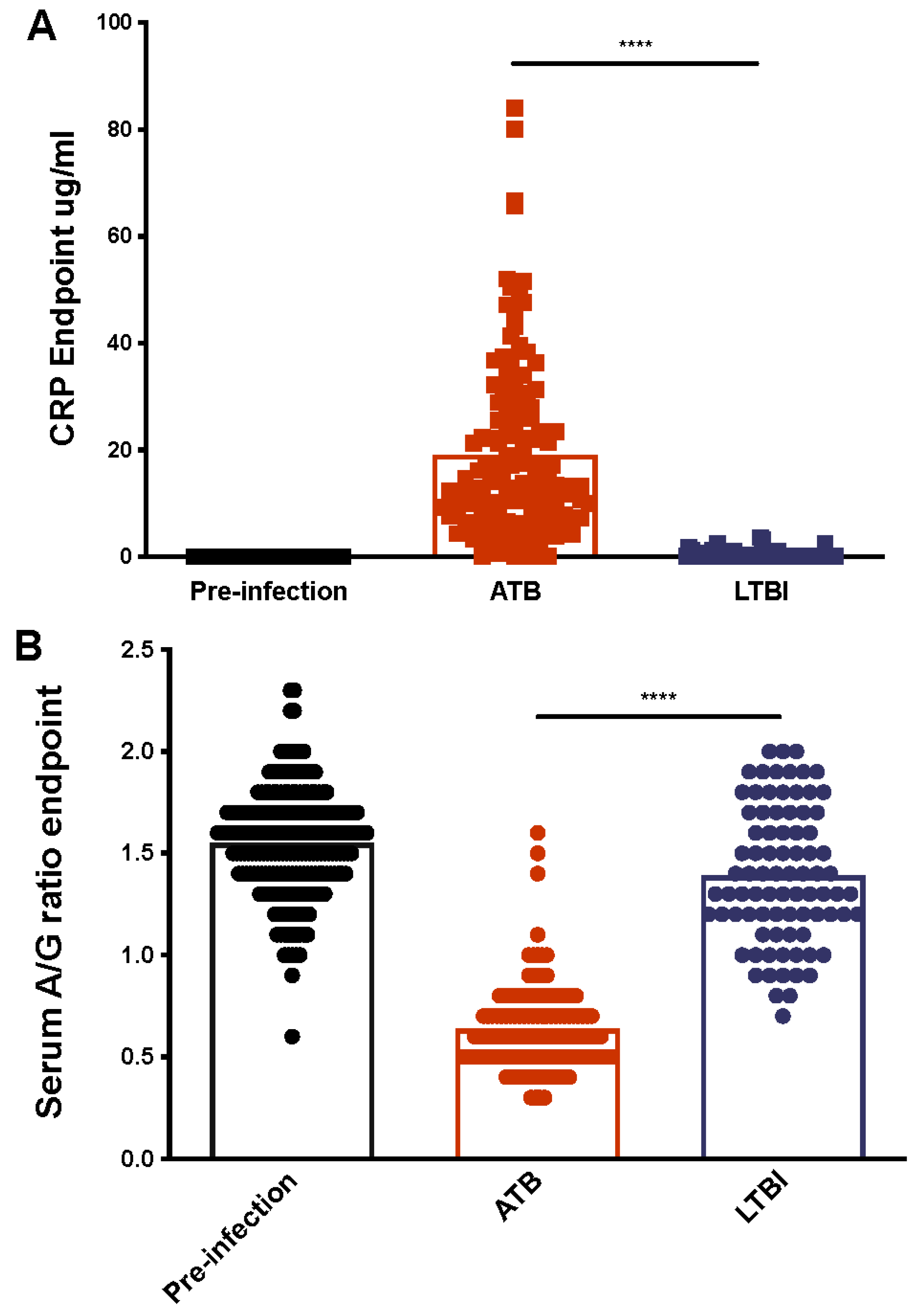
2.2. Serum CRP Levels Compared between ATB and LTBI
2.3. Serum Albumin-to-Globulin (A/G) Ratios Compared between ATB and LTBI
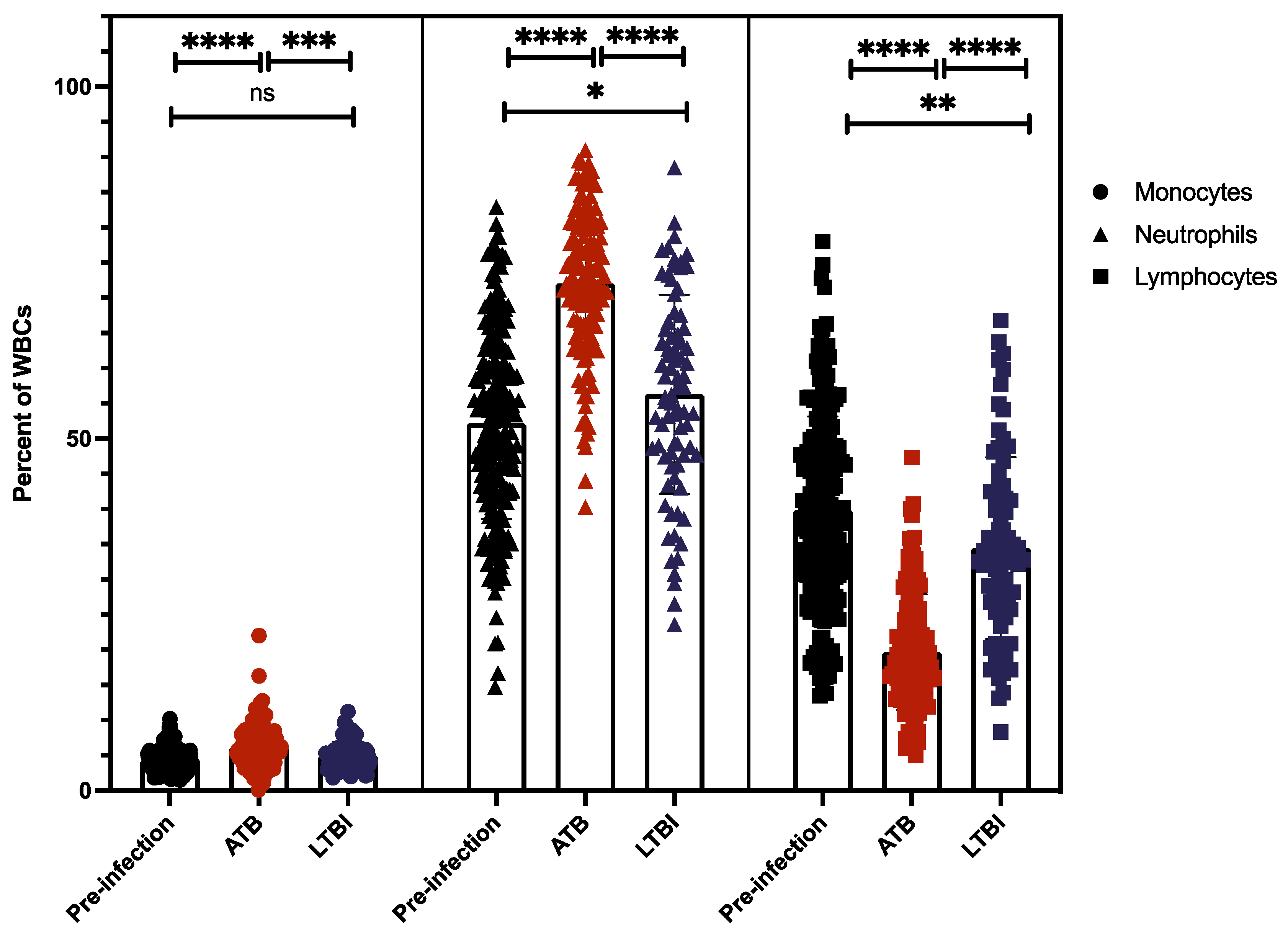
2.4. Myeloid and Lymphoid Cell Counts Compared between ATB and LTBI
3. Methods
3.1. Study Design
3.2. Data Acquisition
3.3. Kynurenine and Tryptophan Ratios in Plasma
3.4. Flow Cytometry
3.5. Difference Analysis
3.6. Correlation
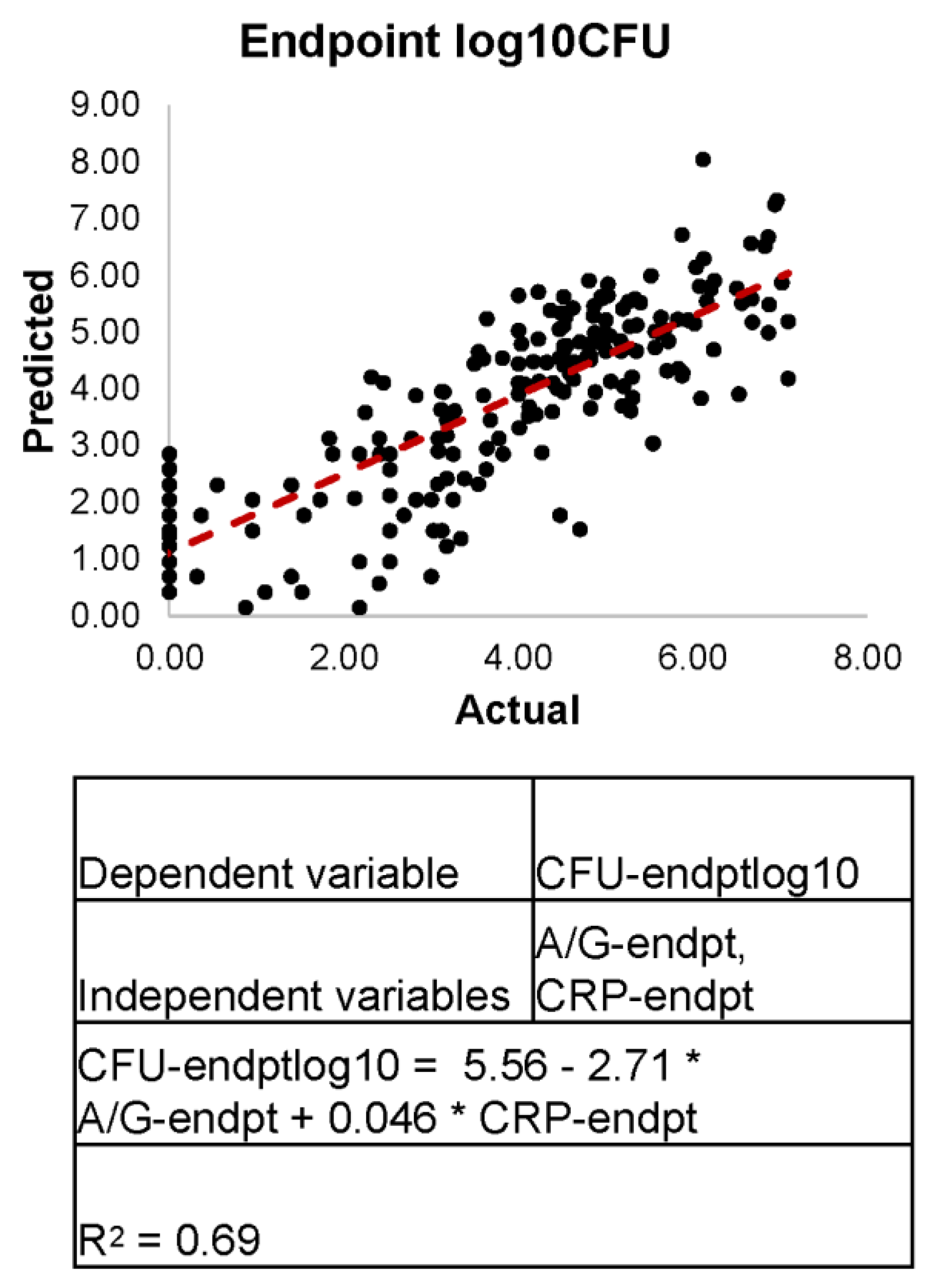
3.7. Linear Regression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2021. Available online: http://www.who.int/tb/publications/global_report/en/ (accessed on 23 April 2019).
- Berry, M.P.R.; Graham, C.M.; McNab, F.W.; Xu, Z.; Bloch, S.A.A.; Oni, T.; Wilkinson, K.A.; Banchereau, R.; Skinner, J.; Wilkinson, R.J.; et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010, 466, 973–977. [Google Scholar] [CrossRef] [PubMed]
- MacLean, E.; Broger, T.; Yerlikaya, S.; Fernandez-Carballo, B.L.; Pai, M.; Denkinger, C.M. A systematic review of biomarkers to detect active tuberculosis. Nat. Microbiol. 2019, 4, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, D.; Mehra, S. Faithful Experimental Models of Human Mycobacterium Tuberculosis Infection. Mycobact. Dis. Tuberc. Lepr. 2012, 2, e108. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, D.; Mehra, S.; Didier, P.J.; Lackner, A.A. The non-human primate model of tuberculosis. J. Med. Primatol. 2012, 41, 191–201. [Google Scholar] [CrossRef]
- Mehra, S.; Foreman, T.W.; Didier, P.J.; Ahsan, M.H.; Hudock, T.A.; Kissee, R.; Golden, N.A.; Gautam, U.S.; Johnson, A.M.; Alvarez, X.; et al. The DosR Regulon Modulates Adaptive Immunity and Is Essential for Mycobacterium tuberculosis Persistence. Am. J. Respir. Crit. Care Med. 2015, 191, 1185–1196. [Google Scholar] [CrossRef]
- Mehra, S.; Golden, N.A.; Dutta, N.K.; Midkiff, C.C.; Alvarez, X.; Doyle, L.A.; Asher, M.; Russell-Lodrigue, K.; Monjure, C.; Roy, C.J.; et al. Reactivation of latent tuberculosis in rhesus macaques by coinfection with simian immunodeficiency virus. J. Med. Primatol. 2011, 40, 233–243. [Google Scholar] [CrossRef]
- Foreman, T.W.; Mehra, S.; LoBato, D.N.; Malek, A.; Alvarez, X.; Golden, N.A.; Bucşan, A.N.; Didier, P.J.; Doyle-Meyers, L.A.; Russell-Lodrigue, K.E.; et al. CD4+ T-cell–independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc. Natl. Acad. Sci. USA 2016, 113, E5636–E5644. [Google Scholar] [CrossRef]
- Phillips, B.L.; Mehra, S.; Ahsan, M.H.; Selman, M.; Khader, S.A.; Kaushal, D. LAG3 Expression in Active Mycobacterium tuberculosis Infections. Am. J. Pathol. 2015, 185, 820–833. [Google Scholar] [CrossRef]
- Danilchanka, O.; Pavlenok, M.; Niederweis, M. Role of Porins for Uptake of Antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 2008, 52, 3127–3134. [Google Scholar] [CrossRef]
- Kaushal, D.; Foreman, T.W.; Gautam, U.S.; Alvarez, X.; Adekambi, T.; Rangel-Moreno, J.; Golden, N.A.; Johnson, A.M.F.; Phillips, B.L.; Ahsan, M.H.; et al. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat. Commun. 2015, 6, 8533. [Google Scholar] [CrossRef]
- Luo, Q.; Mehra, S.; Golden, N.A.; Kaushal, D.; Lacey, M.R. Identification of biomarkers for tuberculosis susceptibility via integrated analysis of gene expression and longitudinal clinical data. Front. Genet. 2014, 5, 240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Levine, D.M.; Dutta, N.K.; Eckels, J.; Scanga, C.; Stein, C.; Mehra, S.; Kaushal, D.; Karakousis, P.C.; Salamon, H. A tuberculosis ontology for host systems biology. Tuberculosis 2015, 95, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Slight, S.R.; Rangel-Moreno, J.; Gopal, R.; Lin, Y.; Junecko, B.A.F.; Mehra, S.; Selman, M.; Becerril-Villanueva, E.; Baquera-Heredia, J.; Pavon, L.; et al. CXCR5+ T helper cells mediate protective immunity against tuberculosis. J. Clin. Investig. 2013, 123, 712–726. [Google Scholar] [CrossRef]
- Darrah, P.A.; Bolton, D.L.; Lackner, A.A.; Kaushal, D.; Aye, P.P.; Mehra, S.; Blanchard, J.L.; Didier, P.J.; Roy, C.J.; Rao, S.S.; et al. Aerosol Vaccination with AERAS-402 Elicits Robust Cellular Immune Responses in the Lungs of Rhesus Macaques but Fails To Protect against High-Dose Mycobacterium tuberculosis Challenge. J. Immunol. 2014, 193, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; McLachlan, J.; Mehra, S.; Kaushal, D. Humoral and lung immune responses to Mycobacterium tuberculosis infection in a primate model of protection. Trials Vaccinol. 2014, 3, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Mehra, S.; Alvarez, X.; Didier, P.J.; Doyle, L.A.; Blanchard, J.L.; Lackner, A.A.; Kaushal, D. Granuloma Correlates of Protection Against Tuberculosis and Mechanisms of Immune Modulation by Mycobacterium tuberculosis. J. Infect. Dis. 2013, 207, 1115–1127. [Google Scholar] [CrossRef]
- Gopal, R.; Monin, L.; Torres, D.; Slight, S.; Mehra, S.; McKenna, K.C.; Fallert Junecko, B.A.; Reinhart, T.A.; Kolls, J.; Báez-Saldaña, R.; et al. S100A8/A9 Proteins Mediate Neutrophilic Inflammation and Lung Pathology during Tuberculosis. Am. J. Respir. Crit. Care Med. 2013, 188, 1137–1146. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mehra, S.; Martinez, A.N.; Alvarez, X.; Renner, N.A.; Morici, L.A.; Pahar, B.; MacLean, A.G.; Lackner, A.A.; Kaushal, D. The Stress-Response Factor SigH Modulates the Interaction between Mycobacterium tuberculosis and Host Phagocytes. PLoS ONE 2012, 7, e28958. [Google Scholar] [CrossRef]
- Mehra, S.; Pahar, B.; Dutta, N.K.; Conerly, C.N.; Philippi-Falkenstein, K.; Alvarez, X.; Kaushal, D. Transcriptional Reprogramming in Nonhuman Primate (Rhesus Macaque) Tuberculosis Granulomas. PLoS ONE 2010, 5, e12266. [Google Scholar] [CrossRef]
- Hudock, T.A.; Foreman, T.W.; Bandyopadhyay, N.; Gautam, U.S.; Veatch, A.V.; LoBato, D.N.; Gentry, K.M.; Golden, N.A.; Cavigli, A.; Mueller, M.; et al. Hypoxia Sensing and Persistence Genes Are Expressed during the Intragranulomatous Survival of Mycobacterium tuberculosis. Am. J. Respir. Cell Mol. Biol. 2017, 56, 637–647. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mehra, S.; Didier, P.J.; Roy, C.J.; Doyle, L.A.; Alvarez, X.; Ratterree, M.; Be, N.A.; Lamichhane, G.; Jain, S.K.; et al. Genetic Requirements for the Survival of Tubercle Bacilli in Primates. J. Infect. Dis. 2010, 201, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Sproston, N.R.; Ashworth, J.J. Role of C-Reactive Protein at Sites of Inflammation and Infection. Front. Immunol. 2018, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Chaisson, L.H.; Patel, S.M.; Drain, P.K.; Wilson, D.; Cattamanchi, A. Diagnostic accuracy of C-reactive protein for active pulmonary tuberculosis: A systematic review and meta-analysis. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 2017, 21, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Geluk, A.; Corstjens, P. CRP: Tell-tale biomarker or common denominator? Lancet Infect. Dis. 2017, 17, 1225–1227. [Google Scholar] [CrossRef]
- Gautam, U.S.; Foreman, T.W.; Bucsan, A.N.; Veatch, A.V.; Alvarez, X.; Adekambi, T.; Golden, N.A.; Gentry, K.M.; Doyle-Meyers, L.A.; Russell-Lodrigue, K.E.; et al. In Vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2018, 115, E62–E71. [Google Scholar] [CrossRef]
- Foreman, T.W.; Veatch, A.V.; LoBato, D.N.; Didier, P.J.; Doyle-Meyers, L.A.; Russell-Lodrigue, K.E.; Lackner, A.A.; Kousoulas, K.G.; Khader, S.A.; Kaushal, D.; et al. Nonpathologic Infection of Macaques by an Attenuated Mycobacterial Vaccine Is not Reactivated in the Setting of HIV Co-Infection. Am. J. Pathol. 2017, 187, 2811–2820. [Google Scholar] [CrossRef]
- Kuroda, M.J.; Sugimoto, C.; Cai, Y.; Merino, K.M.; Mehra, S.; Araínga, M.; Roy, C.J.; Midkiff, C.C.; Alvarez, X.; Didier, E.S.; et al. High Turnover of Tissue Macrophages Contributes to Tuberculosis Reactivation in Simian Immunodeficiency Virus-Infected Rhesus Macaques. J. Infect. Dis. 2018, 217, 1865–1874. [Google Scholar] [CrossRef]
- Mehra, S.; Golden, N.A.; Stuckey, K.; Didier, P.J.; Doyle, L.A.; Russell-Lodrigue, K.E.; Sugimoto, C.; Hasegawa, A.; Sivasubramani, S.K.; Roy, C.J.; et al. The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J. Infect. Dis. 2012, 205, 1203–1213. [Google Scholar] [CrossRef]
- Wang, C.; Wei, L.-L.; Shi, L.-Y.; Pan, Z.-F.; Yu, X.-M.; Li, T.-Y.; Liu, C.M.; Ping, Z.P.; Jiang, T.T.; Chen, Z.L.; et al. Screening and identification of five serum proteins as novel potential biomarkers for cured pulmonary tuberculosis. Sci. Rep. 2015, 5, 15615. [Google Scholar] [CrossRef]
- Saunders, B.M.; Frank, A.A.; Orme, I.M.; Cooper, A.M. CD4 is required for the development of a protective granulomatous response to pulmonary tuberculosis. Cell. Immunol. 2002, 216, 65–72. [Google Scholar] [CrossRef]
- Naranbhai, V.; Hill, A.V.S.; Abdool Karim, S.S.; Naidoo, K.; Abdool Karim, Q.; Warimwe, G.M.; McShane, H.; Fletcher, H. Ratio of Monocytes to Lymphocytes in Peripheral Blood Identifies Adults at Risk of Incident Tuberculosis Among HIV-Infected Adults Initiating Antiretroviral Therapy. J. Infect. Dis. 2014, 209, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yin, Y.; Wang, X.; Pei, H.; Kuai, S.; Gu, L.; Xing, H.; Zhang, Y.; Huang, Q.; Guan, B. Ratio of monocytes to lymphocytes in peripheral blood in patients diagnosed with active tuberculosis. Braz. J. Infect. Dis. 2015, 19, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, R.; Piyaworawong, S.; Naranbhai, V.; Prachamat, P.; Kriengwatanapong, P.; Tsuchiya, N.; Wongyai, J.; Bupachat, S.; Yamada, N.; Summanapan, S.; et al. Predicting the risk of pulmonary tuberculosis based on the neutrophil-to-lymphocyte ratio at TB screening in HIV-infected individuals. BMC Infect. Dis. 2019, 19, 667. [Google Scholar] [CrossRef] [PubMed]
- Mayito, J.; Meya, D.B.; Rhein, J.; Sekaggya-Wiltshire, C. Utility of the monocyte to lymphocyte ratio in diagnosing latent tuberculosis among HIV-infected individuals with a negative tuberculosis symptom screen. PLoS ONE 2020, 15, e0241786. [Google Scholar] [CrossRef]
- Rakotosamimanana, N.; Richard, V.; Raharimanga, V.; Gicquel, B.; Doherty, T.M.; Zumla, A.; Razanamparany, V.R. Biomarkers for risk of developing active tuberculosis in contacts of TB patients: A prospective cohort study. Eur. Respir. J. 2015, 46, 1095–1103. [Google Scholar] [CrossRef]
- Collins, J.M.; Siddiqa, A.; Jones, D.P.; Liu, K.; Kempker, R.R.; Nizam, A.; Shah, N.S.; Ismail, N.; Ouma, S.G.; Tukvadze, N.; et al. Tryptophan catabolism reflects disease activity in human tuberculosis. JCI Insight 2020, 5, e137131. [Google Scholar] [CrossRef]
- Wu, H.; Gong, J.; Liu, Y. Indoleamine 2, 3-dioxygenase regulation of immune response (Review). Mol. Med. Rep. 2018, 17, 4867–4873. [Google Scholar] [CrossRef]
- Thomas, S.M.; Garrity, L.F.; Brandt, C.R.; Schobert, C.S.; Feng, G.S.; Taylor, M.W.; Carlin, J.M.; Byrne, G.I. IFN-gamma-mediated antimicrobial response. Indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. J. Immunol. 1993, 150, 5529–5534. [Google Scholar]
- Zhang, Y.J.; Reddy, M.C.; Ioerger, T.R.; Rothchild, A.C.; Dartois, V.; Schuster, B.M.; Trauner, A.; Wallis, D.; Galaviz, S.; Huttenhower, C.; et al. Tryptophan biosynthesis protects mycobacteria from CD4 T cell-mediated killing. Cell 2013, 155, 1296–1308. [Google Scholar] [CrossRef]
- Esaulova, E.; Das, S.; Singh, D.K.; Choreño-Parra, J.A.; Swain, A.; Arthur, L.; Rangel-Moreno, J.; Ahmed, M.; Singh, B.; Gupta, A.; et al. The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 2021, 29, 165–178.e8. [Google Scholar] [CrossRef]
- Adu-Gyamfi, C.G.; Snyman, T.; Makhathini, L.; Otwombe, K.; Darboe, F.; Penn-Nicholson, A.; Fisher, M.; Savulescu, D.; Hoffmann, C.; Chaisson, R.; et al. Diagnostic accuracy of plasma kynurenine/tryptophan ratio, measured by enzyme-linked immunosorbent assay, for pulmonary tuberculosis. Int. J. Infect. Dis. 2020, 99, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Delemarre, E.M.; van Hoorn, L.; Bossink, A.W.; Drylewicz, J.; Joosten, S.A.; Ottenhoff, T.H.; Akkerman, O.W.; Goletti, D.; Petruccioli, E.; Navarra, A.; et al. Serum Biomarker Profile Including CCL1, CXCL10, VEGF, and Adenosine Deaminase Activity Distinguishes Active From Remotely Acquired Latent Tuberculosis. Front. Immunol. 2021, 12, 725447. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, Z.A.; Wong, E.B.; Ndung'u, T.; Kasprowicz, V.O.; Bishai, W.R. Latent and Active Tuberculosis Infection Increase Immune Activation in Individuals Co-Infected with HIV. eBioMedicine 2015, 2, 334–340. [Google Scholar] [CrossRef]
- Shingdang, J.; Bot, Y.; Ojo, O.I.; Edeh, O.; Essien, C.; Bwende, E.; Okolie, C.; Ekwempu, A.I. Serum Albumin/Globulin ratio in Tuberculosis and HIV Patients any Relationship? Mycobact. Dis. 2016, 6, 199. [Google Scholar]
- Popov, A.; Abdullah, Z.; Wickenhauser, C.; Saric, T.; Driesen, J.; Hanisch, F.G.; Domann, E.; Raven, E.L.; Dehus, O.; Hermann, C.; et al. Indoleamine 2,3-dioxygenase-expressing dendritic cells form suppurative granulomas following Listeria monocytogenes infection. J. Clin. Investig. 2006, 116, 3160–3170. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, P. On watching the watchers: IDO and type I/II IFN. Eur. J. Immunol. 2007, 37, 876–879. [Google Scholar] [CrossRef]
- Wong, C.T.; Saha, N. Serum immunoglobulin and acute phase protein concentrations in pulmonary tuberculosis patients in Singapore. Trop. Geogr. Med. 1989, 41, 218–221. [Google Scholar] [PubMed]
- Dorhoi, A.; Du Plessis, N. Monocytic Myeloid-Derived Suppressor Cells in Chronic Infections. Front. Immunol. 2017, 8, 1895. [Google Scholar] [CrossRef]
- Du Plessis, N.; Loebenberg, L.; Kriel, M.; von Groote-Bidlingmaier, F.; Ribechini, E.; Loxton, A.G.; van Helden, P.D.; Lutz, M.B.; Walzl, G. Increased frequency of myeloid-derived suppressor cells during active tuberculosis and after recent mycobacterium tuberculosis infection suppresses T-cell function. Am. J. Respir. Crit. Care Med. 2013, 188, 724–732. [Google Scholar] [CrossRef]
- McCaffrey, E.F.; Donato, M.; Keren, L.; Chen, Z.; Delmastro, A.; Fitzpatrick, M.B.; Gupta, S.; Greenwald, N.F.; Baranski, A.; Graf, W.; et al. The immunoregulatory landscape of human tuberculosis granulomas. Nat. Immunol. 2022, 23, 318–329. [Google Scholar] [CrossRef]
- Hornyák, L.; Dobos, N.; Koncz, G.; Karányi, Z.; Páll, D.; Szabó, Z.; Halmos, G.; Székvölgyi, L. The Role of Indoleamine-2,3-Dioxygenase in Cancer Development, Diagnostics, and Therapy. Front. Immunol. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Ganatra, S.R.; Bucşan, A.N.; Alvarez, X.; Kumar, S.; Chatterjee, A.; Quezada, M.; Fish, A.; Singh, D.K.; Singh, B.; Sharan, R.; et al. Anti-retroviral therapy does not reduce tuberculosis reactivation in a tuberculosis-HIV co-infection model. J. Clin. Investig. 2020, 130, 5171–5179. [Google Scholar] [CrossRef] [PubMed]
- Sharan, R.; Ganatra, S.R.; Bucsan, A.N.; Cole, J.; Singh, D.K.; Alvarez, X.; Gough, M.; Alvarez, C.; Blakley, A.; Ferdin, J.; et al. Antiretroviral therapy timing impacts latent tuberculosis infection reactivation in a tuberculosis/simian immunodeficiency virus coinfection model. J. Clin. Investig. 2021, 132, e153090. [Google Scholar] [CrossRef] [PubMed]

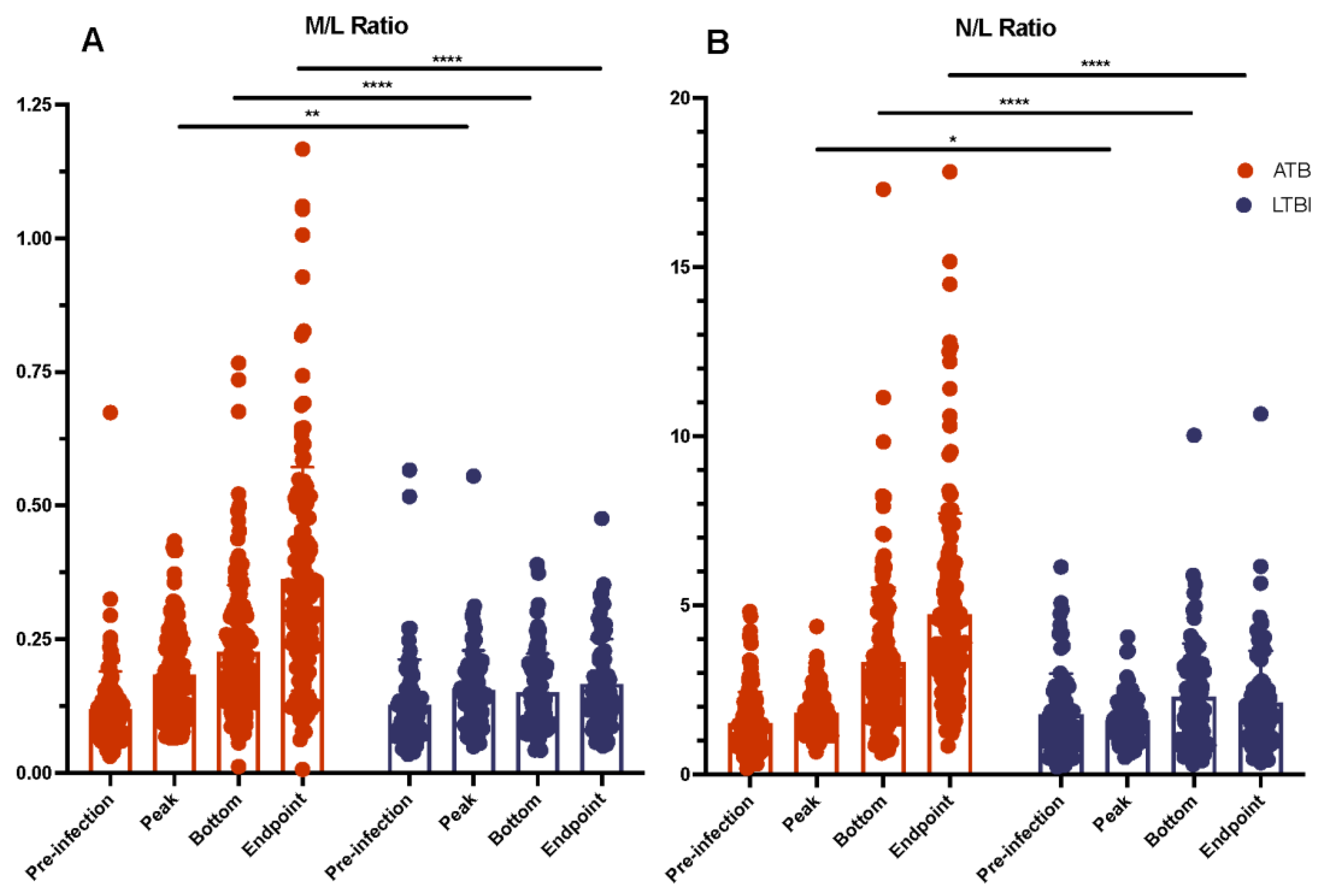

| Welch’s t Test ATB v. LTBI | |
|---|---|
| p-Value | |
| CFU | 4.48 × 10−06 |
| CFU-log10. | 3.31 × 10−44 |
| CRP-pre | |
| CRP-peak | 2.30 × 10−29 |
| CRP-endpt | 6.99 10−27 |
| A/G-pre | |
| A/G-peak | 0.001 |
| A/G-bottom | 5.43 × 10−28 |
| A/G-endpt | 3.43 × 10−36 |
| Lympho-pre | |
| Lympho-peak | |
| Lympho-bottom | 2.44 × 10−07 |
| Lympho-endpt | 2.40 × 10−15 |
| Mono-pre | |
| Mono-peak | |
| Mono-bottom | 0.045 |
| Mono-endpt | 0.001 |
| Neutro-pre | |
| Neutro-peak | 3.38 × 10−06 |
| Neutro-bottom | |
| Neutro-endpt | 2.03 × 10−14 |
| L/M-pre | |
| L/M-peak | 0.012 |
| L/M-bottom | 0.004 |
| L/M-endpt | 0.013 |
| N/M-pre | |
| N/M-peak | |
| N/M-bottom | |
| N/M-endpt | |
| N/L-pre | |
| N/L-peak | 0.021 |
| N/L-bottom | 9.27 × 10−05 |
| N/L-endpt | 2.02 × 10−15 |
| M/L-pre | |
| M/L-peak | 0.007 |
| M/L-bottom | 2.48 × 10−08 |
| M/L-endpt | 8.62 × 10−19 |
| Kynurenine | |
| Tryptophan | 2.82 × 10−04 |
| K/T | 9.21 × 10−04 |
| A. ATB CFU Correlations | B. ATB log10CFU Correlations | ||||
|---|---|---|---|---|---|
| p-Value | Coefficient | p-Value | Coefficient | ||
| CFU-endpt | #N/A | #N/A | CFU-endpt | 0.000 | 0.671 |
| CFU-endptlog10 | 0.000 | 0.671 | CFU-endptlog10 | #N/A | #N/A |
| CRP-pre | CRP-pre | 0.004 | −0.243 | ||
| CRP-peak | 1.43 × 10−08 | 0.458 | CRP-peak | 2.17 × 10−12 | 0.549 |
| CRP-endpt | 2.72 × 10−08 | 0.450 | CRP-endpt | 9.66 × 10−14 | 0.576 |
| A/G-bottom | A/G-bottom | 0.024 | −0.192 | ||
| A/G-endpt | A/G-endpt | 0.003 | −0.250 | ||
| Lympho-bottom | 0.014 | −0.209 | Lympho-bottom | 0.014 | −0.208 |
| Lympho-endpt | 0.013 | −0.212 | Lympho-endpt | ||
| Mono-peak | Mono-peak | ||||
| Mono-endpt | Mono-endpt | ||||
| Neutro-peak | 0.025 | 0.191 | Neutro-peak | ||
| Neutro-endpt | 0.012 | 0.213 | Neutro-endpt | ||
| LM-endpt | 0.037 | 0.178 | LM-endpt | ||
| NM-bottom | 0.029 | 0.186 | NM-bottom | ||
| NM-endpt | 0.012 | 0.212 | NM-endpt | ||
| ML-pre | 0.036 | 0.178 | ML-pre | 0.045 | 0.170 |
| kyn_ng_ml | kyn_ng_ml | 0.014 | 0.453 | ||
| K_T_ng_ug | K_T_ng_ug | 0.027 | 0.411 | ||
| kyn_ug_ml | kyn_ug_ml | 0.014 | 0.453 | ||
| K_T | K_T | 0.027 | 0.411 | ||
| Lung.AM | Lung.AM | ||||
| A. LTBI CFU Correlations | B. LTBI log10CFU Correlations | ||||
|---|---|---|---|---|---|
| p-Value | Coefficient | p-Value | Coefficient | ||
| CFU-endpt | #N/A | #N/A | CFU-endpt | 1.26 × 10−13 | 0.703 |
| CFU-endptlog10 | 1.26 × 10−13 | 0.703 | CFU-endptlog10 | #N/A | #N/A |
| CRP-pre | 0.044 | 0.224 | CRP-pre | ||
| CRP-peak | 0.024 | 0.251 | CRP-peak | 0.001 | 0.365 |
| CRP-endpt | 0.025 | 0.248 | CRP-endpt | 0.028 | 0.244 |
| A/G-bottom | A/G-bottom | 0.015 | −0.268 | ||
| A/G-endpt | 4.89 × 10−04 | −0.377 | A/G-endpt | 3.90 × 10−04 | −0.383 |
| Lympho-bottom | Lympho-bottom | ||||
| Lympho-endpt | Lympho-endpt | ||||
| Mono-peak | 0.034 | 0.234 | Mono-peak | ||
| Mono-endpt | 0.050 | 0.217 | Mono-endpt | ||
| Neutro-peak | Neutro-peak | ||||
| Neutro-endpt | Neutro-endpt | ||||
| LM-endpt | LM-endpt | ||||
| NM-bottom | NM-bottom | ||||
| NM-endpt | NM-endpt | ||||
| ML-pre | ML-pre | ||||
| kyn_ng_ml | kyn_ng_ml | ||||
| K_T_ng_ug | 0.043 | 0.385 | K_T_ng_ug | 0.010 | 0.480 |
| kyn_ug_ml | kyn_ug_ml | ||||
| K_T | 0.043 | 0.385 | K_T | 0.010 | 0.480 |
| Lung.AM | Lung.AM | 0.004 | −0.978 | ||
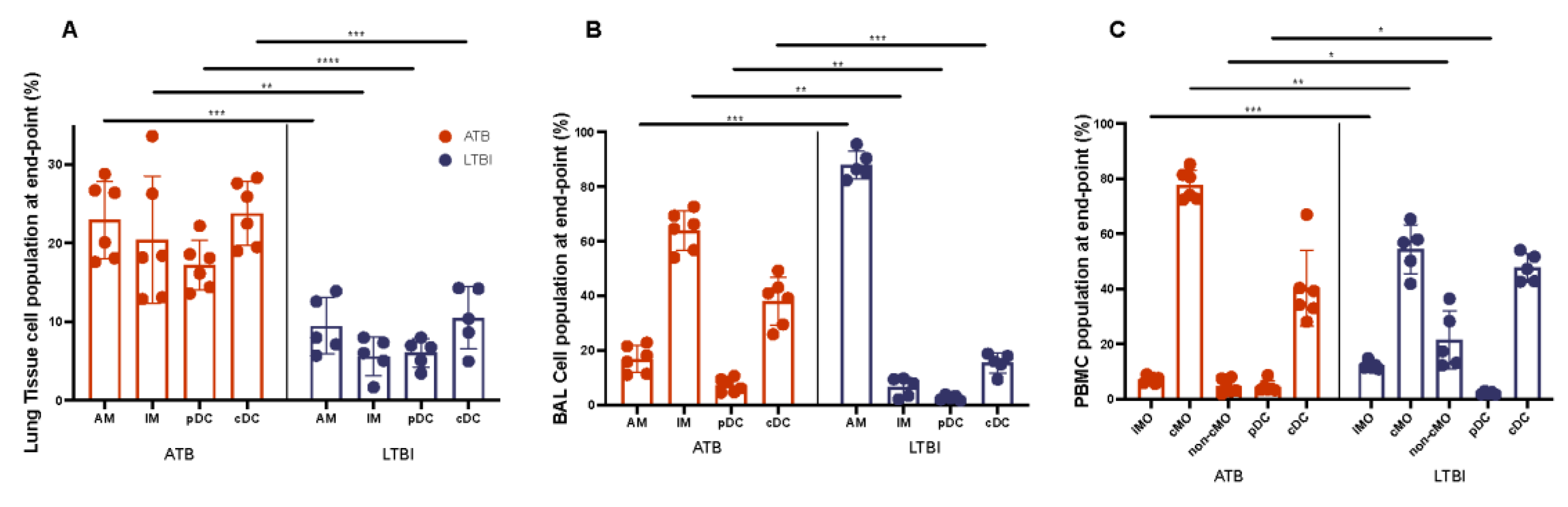
| Welch’s T Test ATB v. LTBI | |||
|---|---|---|---|
| p-Value | |||
| Lung | BAL | PBMC | |
| AM | 5.44 × 10−04 | 1.35 × 10−04 | |
| IM | 5.23 × 10−03 | 3.27 × 10−03 | |
| pDC | 7.58 × 10−05 | 5.26 × 10−03 | 3.99 × 10−02 |
| cDC | 4.24 × 10−04 | 6.99 × 10−04 | |
| iMO | 5.32 × 10−04 | ||
| cMO | 1.79 × 10−03 | ||
| non-cMO | 2.14 × 10−02 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gough, M.; Singh, D.K.; Moodley, C.; Niu, T.; Golden, N.A.; Kaushal, D.; Mehra, S. Peripheral Blood Markers Correlate with the Progression of Active Tuberculosis Relative to Latent Control of Mycobacterium tuberculosis Infection in Macaques. Pathogens 2022, 11, 544. https://doi.org/10.3390/pathogens11050544
Gough M, Singh DK, Moodley C, Niu T, Golden NA, Kaushal D, Mehra S. Peripheral Blood Markers Correlate with the Progression of Active Tuberculosis Relative to Latent Control of Mycobacterium tuberculosis Infection in Macaques. Pathogens. 2022; 11(5):544. https://doi.org/10.3390/pathogens11050544
Chicago/Turabian StyleGough, Maya, Dhiraj K. Singh, Chivonne Moodley, Tianhua Niu, Nadia A. Golden, Deepak Kaushal, and Smriti Mehra. 2022. "Peripheral Blood Markers Correlate with the Progression of Active Tuberculosis Relative to Latent Control of Mycobacterium tuberculosis Infection in Macaques" Pathogens 11, no. 5: 544. https://doi.org/10.3390/pathogens11050544
APA StyleGough, M., Singh, D. K., Moodley, C., Niu, T., Golden, N. A., Kaushal, D., & Mehra, S. (2022). Peripheral Blood Markers Correlate with the Progression of Active Tuberculosis Relative to Latent Control of Mycobacterium tuberculosis Infection in Macaques. Pathogens, 11(5), 544. https://doi.org/10.3390/pathogens11050544






