Immune Responses Regulated by Key Periodontal Bacteria in Germ-Free Mice
Abstract
:1. Introduction
2. Results
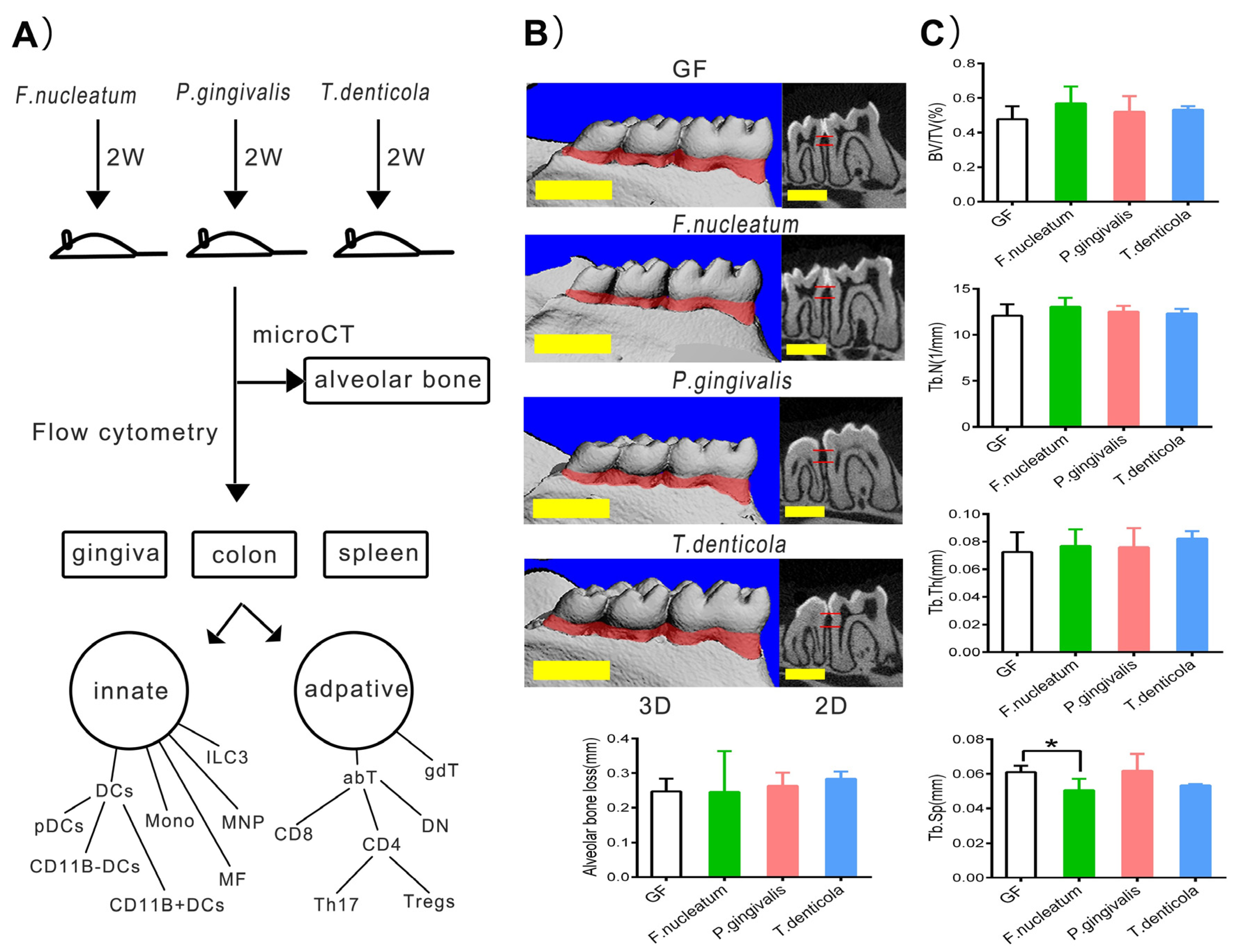
2.1. Bone Resorption after Inoculation with Periodontal Bacteria in Germ-Free Mice
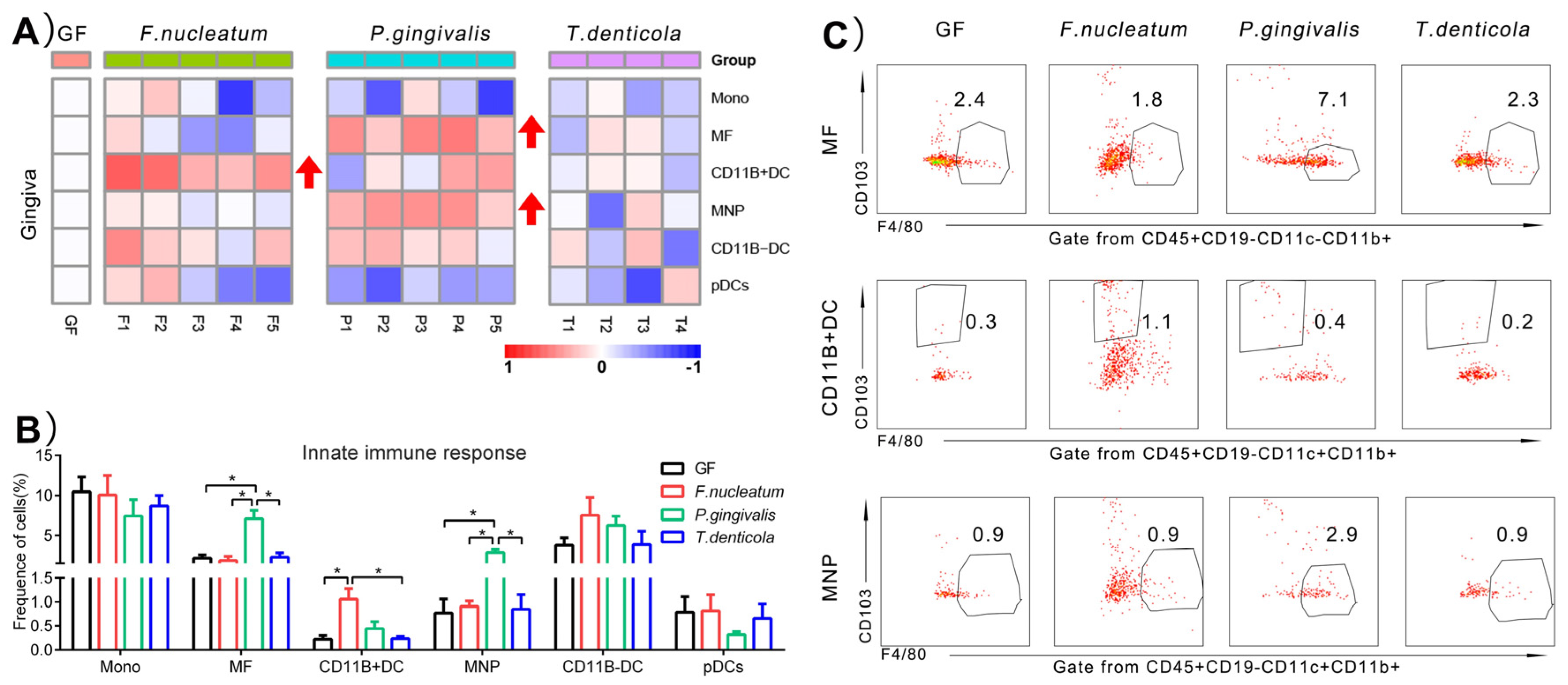
2.2. Immunological Changes of Gingiva in Response to Periodontal Bacteria Colonization
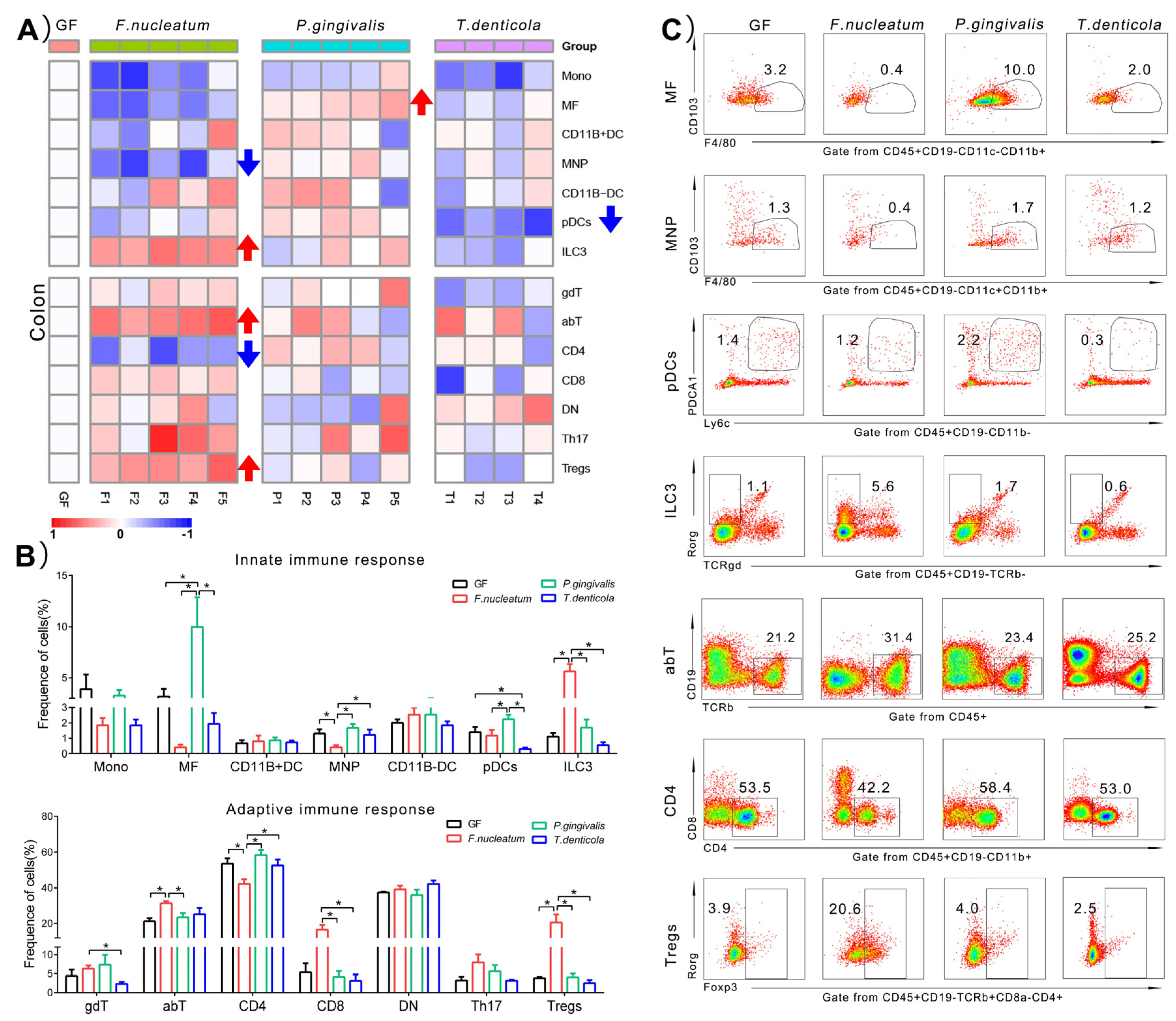
2.3. Immunological Changes of Colon in Response to Periodontal Bacteria Colonization
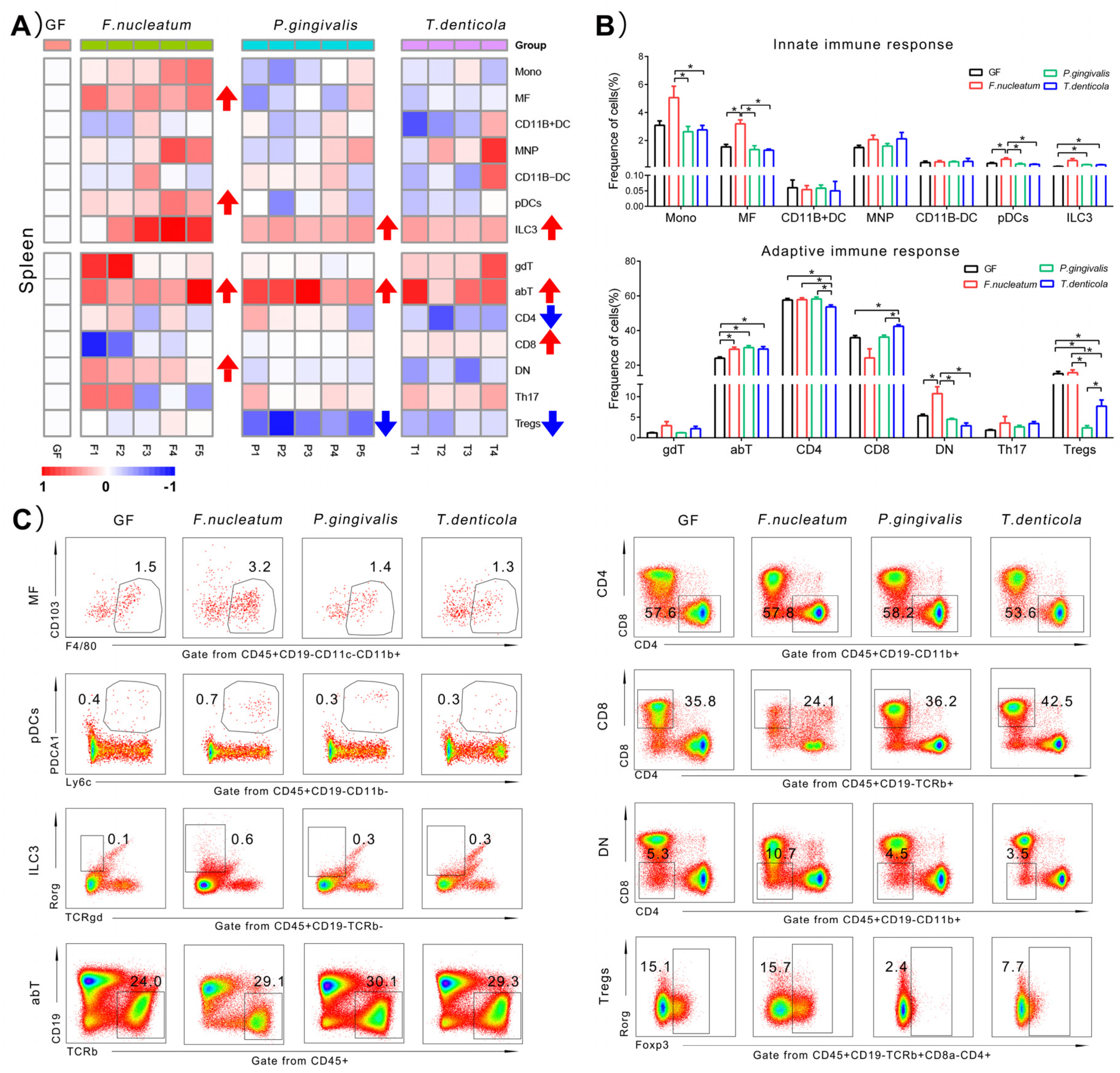
2.4. Immunological Changes of Spleen in Response to Periodontal Bacteria Colonization
2.5. Summary of the Significantly Changing Immune Cell Types
3. Discussion
3.1. P. gingivalis Induced a Higher Innate Immune Response in the Gingiva, Compared to F. nucleatum and T. denticola
3.2. The Local Immunity of the Colon Induced by F. nucleatum Was More Responsive Than P. gingivalis and T. denticola
3.3. One of the Main Effects Induced by T. denticola Was on the Systemic Immune Response
3.4. The Key Immune Cells in Response to Mono-Colonization of Key Periodontal Bacteria
3.5. Direction for Future Research
4. Materials and Methods
4.1. Bacteria and Mice
4.2. Animal Model
4.3. Micro-CT Scanning of Alveolar Bone Resorption
4.4. Immune Cell Preparation and Multi-Color Flow Cytometry Analysis
4.5. Drawing of Heatmap
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, W.; Wang, H.; Liang, S. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 2020, 120, 45–84. [Google Scholar] [CrossRef]
- de Andrade, K.Q.; Almeida-Da-Silva, C.L.C.; Coutinho-Silva, R. Immunological Pathways Triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: Therapeutic Possibilities? Mediat. Inflamm. 2019, 2019, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.; Roger, P.-M. From Crosstalk between Immune and Bone Cells to Bone Erosion in Infection. Int. J. Mol. Sci. 2019, 20, 5154. [Google Scholar] [CrossRef] [Green Version]
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef]
- Tsukasaki, M. RANKL and osteoimmunology in periodontitis. J. Bone Miner. Metab. 2021, 39, 82–90. [Google Scholar] [CrossRef]
- Hickey, N.; Shalamanova, L.; Whitehead, K.A.; Dempsey-Hibbert, N.; Van Der Gast, C.; Taylor, R.L. Exploring the puta-tive interactions between chronic kidney disease and chronic periodontitis. Crit. Rev. Microbiol. 2020, 46, 61–77. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH 1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef]
- Panda, S.K.; Colonna, M. Innate Lymphoid Cells in Mucosal Immunity. Front. Immunol. 2019, 10, 861. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhang, Z.; Wang, Z.-M. Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC Oral Health 2020, 20, 293. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; He, J.; He, Z.; Zhou, Y.; Yuan, M.; Xu, X.; Sun, F.; Liu, C.; Li, J.; Xie, W.; et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014, 8, 1879–1891. [Google Scholar] [CrossRef] [Green Version]
- Hajishengallis, G. The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol. 2014, 29, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Bostanci, N.; Belibasakis, G.N. Porphyromonas gingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 2012, 333, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Dagnæs-Hansen, F.; Løvschall, H.; Song, W.; Nielsen, G.K.; Yang, C.; Wang, Q.; Kjems, J.; Gao, S. Macrophage-mediated nanoparticle delivery to the periodontal lesions in established murine model via Pg-LPS induction. J. Oral Pathol. Med. 2015, 44, 538–542. [Google Scholar] [CrossRef]
- Huang, Y.; Tian, C.; Li, Q.; Xu, Q. TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells. Int. J. Mol. Sci. 2019, 20, 2023. [Google Scholar] [CrossRef] [Green Version]
- Sanders, T.J.; Yrlid, U.; Maloy, K.J. Intestinal Mononuclear Phagocytes in Health and Disease. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Sharawi, H.; Heyman, O.; Mizraji, G.; Horev, Y.; Laviv, A.; Shapira, L.; Yona, S.; Hovav, A.; Wilensky, A. The Prevalence of Gingival Dendritic Cell Subsets in Periodontal Patients. J. Dent. Res. 2021, 100, 1330–1336. [Google Scholar] [CrossRef]
- Goradel, N.H.; Heidarzadeh, S.; Jahangiri, S.; Farhood, B.; Mortezaee, K.; Khanlarkhani, N.; Negahdari, B. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell. Physiol. 2018, 234, 2337–2344. [Google Scholar] [CrossRef]
- Li, B.; Ge, Y.; Cheng, L.; Zeng, B.; Yu, J.; Peng, X.; Zhao, J.; Li, W.; Ren, B.; Li, M.; et al. Oral bacteria colonize and compete with gut microbiota in gnotobiotic mice. Int. J. Oral Sci. 2019, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.-L.; Sztajer, H.; Jarek, M.; Bhuju, S.; Wagner-Döbler, I. Worlds Apart—Transcriptome Profiles of Key Oral Microbes in the Periodontal Pocket Compared to Single Laboratory Culture Reflect Synergistic Interactions. Front. Microbiol. 2018, 9, 124. [Google Scholar] [CrossRef] [Green Version]
- Yousefi, L.; Leylabadlo, H.E.; Pourlak, T.; Eslami, H.; Taghizadeh, S.; Ganbarov, K.; Yousefi, M.; Tanomand, A.; Yousefi, B.; Kafil, H.S. Oral spirochetes: Pathogenic mechanisms in periodontal disease. Microb. Pathog. 2020, 144, 104193. [Google Scholar] [CrossRef]
- Foschi, F.; Izard, J.; Sasaki, H.; Sambri, V.; Prati, C.; Müller, R.; Stashenko, P. Treponema denticola in Disseminating Endodontic Infections. J. Dent. Res. 2006, 85, 761–765. [Google Scholar] [CrossRef] [Green Version]
- Sela, M.N. Role of Treponema Denticola in Periodontal Diseases. Crit. Rev. Oral Biol. Med. 2001, 12, 399–413. [Google Scholar] [CrossRef]
- Jones, M.M.; Vanyo, S.T.; Visser, M.B. The Msp Protein of Treponema denticola Interrupts Activity of Phosphoinositide Processing in Neutrophils. Infect. Immun. 2019, 87, e00553-19. [Google Scholar] [CrossRef] [Green Version]
- Talbot, J.; Hahn, P.; Kroehling, L.; Nguyen, H.; Li, D.; Littman, D.R. Feeding-dependent VIP neuron-ILC3 circui-t regulates the intestinal barrier. Nat. 2020, 579, 575–580. [Google Scholar] [CrossRef]
- Li, C.; Liu, J.; Pan, J.; Wang, Y.; Shen, L.; Xu, Y. ILC1s and ILC3s Exhibit Inflammatory Phenotype in Periodontal Ligament of Periodontitis Patients. Front. Immunol. 2021, 12, 708678. [Google Scholar] [CrossRef]
- Shao, Q.; Gu, J.; Zhou, J.; Wang, Q.; Li, X.; Deng, Z.; Lu, L. Tissue Tregs and Maintenance of Tissue Homeostasis. Front. Cell Dev. Biol. 2021, 9, 717903. [Google Scholar] [CrossRef]
- Izcue, A.; Coombes, J.L.; Powrie, F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol. Rev. 2006, 212, 256–271. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Jin, Y.; Gao, H.; Lin, X. Oral Administration of All-Trans Retinoic Acid Suppresses Experimental Periodontitis by Modulating the Th17/Treg Imbalance. J. Periodontol. 2014, 85, 740–750. [Google Scholar] [CrossRef]
- Tang, Z.; Cheng, X.; Su, X.; Wu, L.; Cai, Q.; Wu, H. Treponema denticola Induces Alzheimer-Like Tau Hyperphosphorylation by Activating Hippocampal Neuroinflammation in Mice. J. Dent. Res. 2022, 220345221076772. [Google Scholar] [CrossRef]
- Wilharm, A.; Tabib, Y.; Nassar, M.; Reinhardt, A.; Mizraji, G.; Sandrock, I.; Heyman, O.; Barros-Martins, J.; Aizenbud, Y.; Khalaileh, A.; et al. Mutual interplay between IL-17–producing γδT cells and microbiota orchestrates oral mucosal homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 2652–2661. [Google Scholar] [CrossRef] [Green Version]
| Organs | F. nucleatum | P. gingivalis | T. denticola |
|---|---|---|---|
| Gingiva | CD11B+DC↑ | MNPs↑, MFs↑ | / |
| Colon | ILC3s↑, MNPs↓ Tregs↑, abT↑ CD4+T↓ | MFs↑ | pDCs↓ |
| Spleen | MFs↑, pDCs↑ DN↑, abT↑ | ILC3s↑ Tregs↓, abT↑ | ILC3s↑ abT↑, CD8+T↓ CD4+T↓, Tregs↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Yang, Y.; Li, J.; Zhang, B.; Wei, W.; Lu, C.; Yan, C.; Wei, H.; Li, Y. Immune Responses Regulated by Key Periodontal Bacteria in Germ-Free Mice. Pathogens 2022, 11, 513. https://doi.org/10.3390/pathogens11050513
Shen X, Yang Y, Li J, Zhang B, Wei W, Lu C, Yan C, Wei H, Li Y. Immune Responses Regulated by Key Periodontal Bacteria in Germ-Free Mice. Pathogens. 2022; 11(5):513. https://doi.org/10.3390/pathogens11050513
Chicago/Turabian StyleShen, Xin, Yutao Yang, Jian Li, Bo Zhang, Wei Wei, Changqing Lu, Caixia Yan, Hong Wei, and Yan Li. 2022. "Immune Responses Regulated by Key Periodontal Bacteria in Germ-Free Mice" Pathogens 11, no. 5: 513. https://doi.org/10.3390/pathogens11050513
APA StyleShen, X., Yang, Y., Li, J., Zhang, B., Wei, W., Lu, C., Yan, C., Wei, H., & Li, Y. (2022). Immune Responses Regulated by Key Periodontal Bacteria in Germ-Free Mice. Pathogens, 11(5), 513. https://doi.org/10.3390/pathogens11050513





