Abstract
We are interested in identifying and characterizing small molecule inhibitors of bacterial virulence factors for their potential use as anti-virulence inhibitors. We identified from high-throughput screening assays a potential activity for avasimibe, a previously characterized acyl-coenzyme A: cholesterol acyltransferase inhibitor, in inhibiting the NleB and SseK arginine glycosyltransferases from Escherichia coli and Salmonella enterica, respectively. Avasimibe inhibited the activity of the Citrobacter rodentium NleB, E. coli NleB1, and S. enterica SseK1 enzymes, without affecting the activity of the human serine/threonine N-acetylglucosamine (O-GlcNAc) transferase. Avasimibe was not toxic to mammalian cells at up to 200 µM and was neither bacteriostatic nor bactericidal at concentrations of up to 125 µM. Doses of 10 µM avasimibe were sufficient to reduce S. enterica abundance in RAW264.7 macrophage-like cells, and intraperitoneal injection of avasimibe significantly reduced C. rodentium survival in mice, regardless of whether the avasimibe was administered pre- or post-infection. We propose that avasimibe or related derivates created using synthetic chemistry may have utility in preventing or treating bacterial infections by inhibiting arginine glycosyltransferases that are important to virulence.
1. Introduction
We have characterized a conserved group of type III secretion system (T3SS) effector proteins that inhibit innate immune responses to infection [1,2,3,4,5]. These proteins (named NleB in enterohemorrhagic and enteropathogenic Escherichia coli (EHEC and EPEC) and SseK in Salmonella enterica) are glycosyltransferases that are important for bacterial virulence. These enzymes glycosylate host protein substrates with β-D-N-acetylglucosamine (GlcNAc) on arginine residues.
Several “death domain”-containing proteins such as the Fas-associated protein with death domain (FADD), tumor necrosis factor receptor type 1-associated death domain protein (TRADD), and RIPK1 are substrates of the NleB/SseK glycosyltransferases [6]. NleB1 disrupts tumor necrosis factor receptor (TNFR)-associated factor (TRAF) signaling, leading to inhibition of the proinflammatory NF-κB pathway [7]. Another target of NleB1 is glyceraldehyde 3-phosphate dehydrogenase (GAPDH). In addition to its role in glycolysis, GAPDH binds to TRAF proteins and stimulates TRAF polyubiquitination [4].
Arginine glycosylation is biologically important because the glycosylation of arginines on host protein substrates leads to their irreversible inactivation and disrupts the normal functioning of the innate immune response. Mammals do not have the enzymatic machinery to add GlcNAc residues to arginine (N-GlcNAc), while this modification is absolutely critical for E. coli and Salmonella virulence. Inhibitors that prevent the formation of this unusual post-translational modification represent a potentially novel way to combat these infections. Non-traditional antibacterial therapeutic strategies have recently been reviewed and are of emerging interest [8].
We previously developed a high-throughput screening (HTS) assay to identify NleB/SseK inhibitors [9,10]. Here, we show that avasimibe (Figure 1A), an acyl-coenzyme A: cholesterol acyltransferase inhibitor [11], also inhibits NleB and SseK enzyme activity, leading to reduced pathogen colonization in macrophage and mouse models of Salmonella and Citrobacter rodentium infection, respectively.
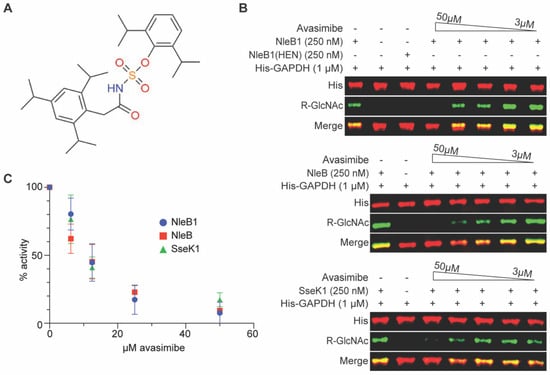
Figure 1.
In vitro glycosylation assays. (A) Avasimibe structure. (B) Western blot results for avasimibe inhibition of NleB1, NleB, and SseK1 (250 nM) glycosylation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 1 µM). Avasimibe was added at concentrations from 3 to 50 µM. The NleB1(HEN) enzyme is an inactive negative control [5]. (C) Quantification of Western blot signal intensities from replicates of in vitro glycosylation assays, n = 3 independent experimental replicates.
2. Results
2.1. Avasimibe Inhibits NleB1 and Its Orthologs
We previously published the results of HTS assays in which we identified several small molecules (100066N, 102644N, and YM155) capable of inhibiting NleB/SseK enzyme activity in vitro [9,10]. In those previous studies, we also discovered a potential activity for avasimibe in inhibiting EHEC NleB1 activity. We first validated the HTS data by quantifying the extent to which avasimibe could prevent Arg-glycosylation of the GAPDH substrate by the EHEC NleB1, Citrobacter NleB, and Salmonella SseK1 enzymes in vitro. We conducted the glycosylation assays in the presence of 2-fold serial dilutions of avasimibe. Avasimibe inhibited each enzyme in a concentration-dependent manner, with apparent IC50s of ~10 µM (Figure 1B,C).
2.2. Avasimibe Does Not Inihibit the Mammalian OGT Enzyme, Is Not Toxic to Mammalian Cells, and Is Neither Bacteriostatic nor Bacteriocidal
We next examined whether avasimibe affected the activity of the human serine/threonine N-acetylglucosamine (O-GlcNAc) transferase (OGT) that regulates protein glycosylation [12]. To assess OGT activity as a function of avasimibe concentration, we used a bioluminescence-based UDP-Glo glycosyltransferase assay. Avasimbe had no effect on human OGT activity in vitro at concentrations of up to 200 µM (Figure 2A). Avasimibe was also found to be nontoxic to mammalian cells at concentrations of up to 50 µM, as determined by conducting a 3-(4,5 dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure 2B). Avasimbe had no effect on the growth rates of C. rodentium or Salmonella enterica at concentrations of up to 125 µM when these bacteria were grown in LB broth (Figure 2C,D). Thus, avasimibe does not appear to act as a general bacteriostatic or bactericidal agent.
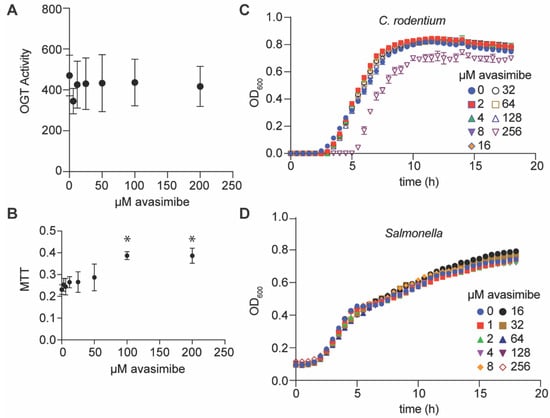
Figure 2.
OGT activity, cytotoxicity, and bacterial growth assays. (A) OGT activity assay. OGT activity was measured by using a UDP-Glo assay in the presence of avasimibe, n = 3 independent experimental replicates. (B) Cell cytotoxicity as measured by performing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Avasimibe was added to RAW264.7 cells for 24 h, and cell viability was assayed by monitoring MTT signal intensity, n = 3 independent experimental replicates. Asterisks (*) indicate significantly different MTT signals as compared to untreated cells, p < 0.05, Dunn’s multiple comparisons test. (C,D) Bacterial growth assays. C. rodentium and S. enterica were cultured in LB media at 37 °C in the presence of the indicated concentrations of avasimibe. Bacterial growth was monitored as a function of time, n = 3 independent experimental replicates.
2.3. Avasimibe Inhibits Salmonella and C. rodentium Survival In Vivo
We quantified the impact of avasimibe on Salmonella survival in cell culture to determine whether it can be used to reduce pathogen loads in mammalian cells. When avasimibe was provided to RAW264.7 cells prior to Salmonella infection at concentrations greater than 10 µM, the amount of intracellular Salmonella was significantly reduced 24 h later (Figure 3A).
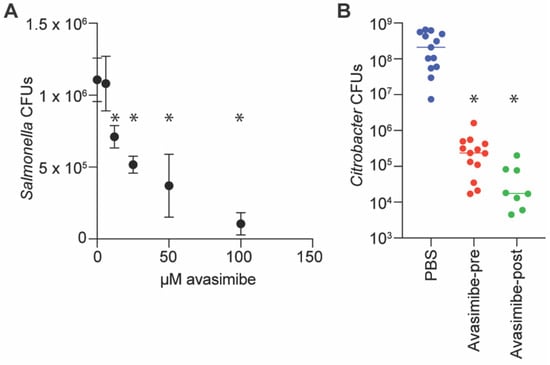
Figure 3.
Infection assays. (A) Salmonella infection assays. RAW264.7 cells were seeded at 1 × 105 cells/well in 24-well plates, and avasimibe was added 1 h before infection with 106 CFUs of Salmonella for 30 min. Cells were incubated in medium containing 100 µg/mL gentamicin for 1 h, and then in 10 µg/mL gentamicin for an additional 23 h. Bacteria were released from RAW264.7 cells using 1% saponin, diluted in PBS, and plated for colony counts, n = 3 independent experimental replicates. Asterisks (*) indicate significantly different Salmonella CFUs as compared to untreated macrophages, p < 0.05, Dunn’s multiple comparisons test. (B) C. rodentium infections of mice. Mice were infected via oral gavage with 109 CFUs of C. rodentium in 100 µL PBS. Avasimibe (25 mg/kg) was administered to one group of mice via intraperitoneal (IP) injection immediately before oral gavage. Avasimibe (5 mg/kg) was provided to another group of mice via IP injection at 24 and 48 h after oral gavage. Asterisks indicate significantly different CFUs as compared to untreated mice, p < 0.05, Dunn’s, n = 8–13 mice.
We performed a mouse infection experiment to determine whether avasimibe inhibition of NleB had any effect in vivo. We administered avasimibe via intraperitoneal injection at doses of either 25 mg/kg immediately prior to infecting mice with C. rodentium, or 5 mg/kg at 24 and 48 h post-infection. After 7 days, we euthanized the mice and quantified the amount of C. rodentium in the colon. We observed a significant reduction in C. rodentium in mice treated with avasimibe, as compared to untreated mice, regardless of whether the avasimibe was administered pre- or post-infection (Figure 3B). These findings suggest that avasimibe could be used as an anti-virulence small molecule.
3. Discussion
We discovered here that avasimibe, a previously characterized ACAT inhibitor, also inhibits the NleB and SseK arginine glycosyltransferases. Avasimibe has a well-documented solubility and safety profile [13], although it caused a potential reduction in the potency of Lipitor [11] and was thus not ultimately used to treat hyperlipidemia or atherosclerosis. Avasimibe also suppresses tumor proliferation and metastasis via the E2F-1 signaling pathway and has potential utility in treating prostate cancer [14]. Avasimibe alleviates insulin resistance in diet-induced obese mice [15]. Avasimbe impedes tick embryo development by interfering with tick lipid metabolism, making ticks more susceptible to bacterial infection [16]. Avasimibe has also shown encouraging results in inhibiting glioma cell proliferation [17]. Additionally, avasimibe was identified as a potential hepatitis C virus inhibitor, where it targets the assembly of infectious viral particles [18].
Our glycosylation assay results suggest that avasimibe has an IC50 of ~10 µM against the NleB/SseK enzymes. At these concentrations, avasimibe has no substantial inhibitory effect on pathogen growth, no cytotoxicity to mammalian cells, and does not inhibit the human OGT enzyme. Finally, our in vivo findings suggest that avasimibe is well tolerated in mice and significantly reduces C. rodentium loads in the intestine, regardless of whether it is administered pre- or post-infection.
Numerous previous investigations have established the safety profile of avasimibe in mice. These studies included prolonged administration of similar doses of avasimibe as those used in our studies, with no reported adverse effects. For example, in a study of Lewis lung carcinoma in mice, 15 mg/kg of avasimibe was administered for 25 days via IP injection with no adverse effects in mice [19]. A similar study used a 7.5 mg/kg dose of avasimibe via IP injection for 30 days and found no adverse effects [20]. A 10 mg/kg dose of avasimibe for up to 22 weeks of daily administration had no effect on body weight or food intake of mice [21].
We have now identified and studied a number of inhibitors that have the potential to function as anti-virulence therapeutics against enteric pathogens. We were indeed surprised to find that avasimibe functions as an inhibitor of bacterial glycosyltransferases. Our future goals are to acquire structural information using NMR and crystallography to characterize the avasimibe binding site and mode of action. Our ultimate goal is to combine our understanding of the molecular mechanisms of action of these inhibitors to create new compounds with improved solubility, lower production costs, and good safety profiles. For example, using synthetic chemistry to modify avasimibe and reduce its interaction with the liver enzyme CYP3A4 [22] could reduce its side effects in humans while maintaining its safety and efficacy profile.
4. Materials and Methods
4.1. Cell Lines
Abelson-murine-leukemia-virus-induced, macrophage-like cells from BALB/c mice (RAW264.7) were purchased from ATCC and grown in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Minneapolis, MN, USA) and 100 µg/mL penicillin/streptomycin (Sigma, St. Louis, MO, USA) in 5% CO2.
4.2. Protein Purification
Protein purification was conducted as previously described [1]. E. coli BL21 (DE3) strains were cultured in LB at 37 °C until an OD600 of 0.4, after which 0.5 mM IPTG was added for 4 h. The pellet was resuspended in 50 mM sodium phosphate buffer pH 8.0 and 0.5 mg/mL lysozyme after centrifugation. After 30 min on ice with periodic shaking, the suspension was treated with 50 mM sodium phosphate buffer pH 8.0, 2 M NaCl, 8 mM imidazole, 20% glycerol, and 2% Triton X-100 for 30 min. After sonicating and centrifuging the cell lysates, the supernatant was added to 2 mL Ni-NTA resin (Qiagen, Germantown, MD, USA) for 1 h of rotation at 4 °C. The mixture was added to a Poly-Prep Chromatography Column (Bio-Rad, Hercules, CA, USA) and washed with 10 mL of 50 mM sodium phosphate buffer pH 8.0, 600 mM NaCl, 60 mM imidazole, and 10% glycerol. Proteins were eluted in 2 mL 50 mM sodium phosphate buffer pH 8.0, 600 mM NaCl, 250 mM imidazole, and 10% glycerol, then dialyzed into the same buffer lacking imidazole.
4.3. Glycosylation Assays
In vitro glycosylation assays were conducted as previously described [1]. Enzymes (200 nM of NleB1, NleB, or SseK1) were incubated in 50 mM Tris-HCl buffer pH 7.4, 1 mM UDP-GlcNAc, 10 mM MnCl2, and 1 mM DTT with 1 mM GAPDH in the presence or absence of serial dilutions of avasimibe. After a 2 h incubation at room temperature, samples were blotted with anti-R-GlcNAc and anti-His tag monoclonal antibodies (Abcam, Cambridge, MA, USA). LI-COR Image Studio software (LI-COR Biosciences, Lincoln, NY, USA) was used to measure signal intensities, and inhibition was estimated by measuring the relative reduction in substrate glycosylation.
4.4. OGT Assays
The UDP-GloTM Glycosyltransferase Assay Kit was used as specified by the manufacturer (Promega). OGT (200 nM) in 25 mM Tris-HCL buffer pH 7.5, 12.5 mM MgCl2, 0.06 mg/mL BSA, 1 mM DTT, 50 µM OGT peptide substrate, and 100 µM UDP-GlcNAc were used in the reactions, which also included avasimibe at concentrations ranging from 6 to 200 µM. The reactions were incubated at 22 °C for 1 h, and the luminescence signals were measured by using a FLUO star microplate reader (BMG Labtech, Cary, NC, USA).
4.5. MTT Assays
MTT tests utilizing RAW264.7 cells in the presence of 2-fold serial dilutions of avasimibe from 3 to 200 µM concentration were performed as specified by Millipore. Formazan absorbance was measured at 570 nm using a BioTek Microplate reader (BioTek, Winooski, VT, USA).
4.6. Bacterial Growth Assays
Bacterial cultures were grown overnight, diluted 1:200 in LB, and then grown at 37 °C for 18 h in the presence of 2-fold serial dilutions of avasimibe (0–256 µM). The absorbance of the culture medium at OD600 was used to monitor bacterial growth.
4.7. Macrophage Infections
RAW264.7 cells were seeded at 1 × 105 cells/well in TCP 24-well tissue culture plates, and avasimibe (6–100 µM) was added 1 h before infection. Salmonella cultures were grown overnight, and 106 CFUs were added to each well for 30 min. Cells were treated with 100 µg/mL gentamicin for 1 h and then with 10 µg/mL gentamicin for an additional 23 h. Bacteria were released from RAW264.7 cells using 1% saponin (Sigma), diluted in PBS, and plated to enumerate the number of intracellular Salmonella.
4.8. Mouse Infections
Five-week-old C57BL/6 mice (Jackson Laboratory) were housed at Kansas State University. C. rodentium DBS100 was cultivated in LB broth with shaking at 200 rpm at 37 °C overnight. Mice were infected via oral gavage with 109 CFUs of C. rodentium in 100 µL PBS. Avasimibe was administered to one group of mice via intraperitoneal (IP) injection immediately before oral gavage. Avasimibe was provided to another group of mice via IP injection at 24 and 48 h after oral gavage. Mice were euthanized 7 days after infection, colons were homogenized, serially diluted, and plated on MacConkey agar. The following day, colonies were enumerated and plotted to compare C. rodentium loads between experimental groups.
Author Contributions
P.R.H. conceived and coordinated the study and wrote the paper. M.K.H., S.E.Q., P.M. and A.R. performed and analyzed the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
The project described was supported by grant number AI153202 from the National Institute of Allergy and Infectious Diseases (NIAID) and by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award numbers P20GM130448, P20GM113117, and P20GM103638, as well as by a CMLD Legacy (GM111385) grant. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID or the National Institute of General Medical Sciences (NIGMS).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of Kansas State University (protocol #454, approved 04/09/2021).
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- El Qaidi, S.; Chen, K.; Halim, A.; Siukstaite, L.; Rueter, C.; Hurtado-Guerrero, R.; Clausen, H.; Hardwidge, P.R. Nleb/Ssek Effectors from Citrobacter Rodentium, Escherichia Coli, and Salmonella Enterica Display Distinct Differences in Host Substrate Specificity. J. Biol. Chem. 2017, 292, 11423–11430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Qaidi, S.; Scott, N.E.; Hays, M.P.; Geisbrecht, B.V.; Watkins, S.; Hardwidge, P.R. An Intra-Bacterial Activity for a T3ss Effector. Sci. Rep. 2020, 10, 1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Pham, T.H.; Feuerbacher, L.A.; Chen, K.; Hays, M.P.; Singh, G.; Rueter, C.; Hurtado-Guerrero, R.; Hardwidge, P.R. Citrobacter Rodentium Nleb Protein Inhibits Tumor Necrosis Factor (Tnf) Receptor-Associated Factor 3 (Traf3) Ubiquitination to Reduce Host Type I Interferon Production. J. Biol. Chem. 2016, 291, 18232–18238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Wang, X.; Pham, T.H.; Feuerbacher, L.A.; Lubos, M.-L.; Huang, M.; Olsen, R.; Mushegian, A.; Slawson, C.; Hardwidge, P.R. Nleb, a Bacterial Effector with Glycosyltransferase Activity, Targets Gapdh Function to Inhibit Nf-Kappab Activation. Cell Host Microbe 2013, 13, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Park, J.B.; Kim, Y.H.; Yoo, Y.; Kim, J.; Jun, S.-H.; Cho, J.W.; El Qaidi, S.; Walpole, S.; Monaco, S.; García-García, A.A.; et al. Structural Basis for Arginine Glycosylation of Host Substrates by Bacterial Effector Proteins. Nat. Commun. 2018, 9, 4283. [Google Scholar] [CrossRef]
- Li, S.; Zhang, L.; Yao, Q.; Li, L.; Dong, N.; Rong, J.; Gao, W.; Ding, X.; Sun, L.; Chen, X.; et al. Pathogen Blocks Host Death Receptor Signalling by Arginine Glcnacylation of Death Domains. Nature 2013, 501, 242–246. [Google Scholar] [CrossRef]
- Pearson, J.S.; Giogha, C.; Ong, S.Y.; Kennedy, C.L.; Kelly, M.; Robinson, K.S.; Lung, T.W.; Mansell, A.; Riedmaier, P.; Oates, C.V.; et al. A Type Iii Effector Antagonizes Death Receptor Signalling During Bacterial Gut Infection. Nature 2013, 501, 247–251. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Piddock, L.J.V. Non-Traditional Antibacterial Therapeutic Options and Challenges. Cell Host Microbe 2019, 26, 61–72. [Google Scholar] [CrossRef]
- El Qaidi, S.; Zhu, C.; McDonald, P.; Roy, A.; Maity, P.K.; Rane, D.; Perera, C.; Hardwidge, P.R. High-Throughput Screening for Bacterial Glycosyltransferase Inhibitors. Front. Cell Infect Microbiol. 2018, 8, 435. [Google Scholar] [CrossRef]
- Zhu, C.; El Qaidi, S.; McDonald, P.; Roy, A.; Hardwidge, P. Ym155 Inhibits Nleb and Ssek Arginine Glycosyltransferase Activity. Pathogens 2021, 10, 253. [Google Scholar] [CrossRef]
- Burnett, J.R.; Huff, M.W. Avasimibe Pfizer. Curr. Opin. Investig. Drugs 2002, 3, 1328–1333. [Google Scholar] [PubMed]
- Aquino-Gil, M.; Pierce, A.; Perez-Cervera, Y.; Zenteno, E.; Lefebvre, T. Ogt: A Short Overview of an Enzyme Standing out from Usual Glycosyltransferases. Biochem. Soc. Trans. 2017, 45, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Burnett, J.R.; Wilcox, L.J.; Telford, D.E.; Kleinstiver, S.J.; Barrett, P.H.; Newton, R.S.; Huff, M.W. Inhibition of Acat by Avasimibe Decreases Both Vldl and Ldl Apolipoprotein B Production in Miniature Pigs. J. Lipid Res. 1999, 40, 1317–1327. [Google Scholar] [CrossRef]
- Xiong, K.; Wang, G.; Peng, T.; Zhou, F.; Chen, S.; Liu, W.; Ju, L.; Xiao, Y.; Qian, K.; Wang, X. The Cholesterol Esterification Inhibitor Avasimibe Suppresses Tumour Proliferation and Metastasis Via the E2f-1 Signalling Pathway in Prostate Cancer. Cancer Cell Int. 2021, 21, 461. [Google Scholar] [CrossRef]
- Zhu, Y.; Kim, S.Q.; Zhang, Y.; Liu, Q.; Kim, K.H. Pharmacological Inhibition of Acyl-Coenzyme A:Cholesterol Acyltransferase Alleviates Obesity and Insulin Resistance in Diet-Induced Obese Mice by Regulating Food Intake. Metabolism 2021, 123, 154861. [Google Scholar] [CrossRef]
- Xavier, M.A.; Brust, F.R.; Waldman, J.; Macedo, A.J.; Juliano, M.A.; da Silva Vaz, I., Jr.; Termignoni, C. Interfering with Cholesterol Metabolism Impairs Tick Embryo Development and Turns Eggs Susceptible to Bacterial Colonization. Ticks Tick Borne Dis. 2021, 12, 101790. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, L.; Li, X.; Shi, Y. Avasimibe Inhibits the Proliferation, Migration and Invasion of Glioma Cells by Suppressing Linc00339. Biomed. Pharmacother. 2020, 130, 110508. [Google Scholar] [CrossRef]
- Hu, L.; Li, J.; Cai, H.; Yao, W.; Xiao, J.; Li, Y.-P.; Qiu, X.; Xia, H.; Peng, T. Avasimibe: A Novel Hepatitis C Virus Inhibitor That Targets the Assembly of Infectious Viral Particles. Antiviral Res. 2017, 148, 5–14. [Google Scholar] [CrossRef]
- Bi, M.; Qiao, X.; Zhang, H.; Wu, H.; Gao, Z.; Zhou, H.; Shi, M.; Wang, Y.; Yang, J.; Hu, J.; et al. Effect of Inhibiting Acat-1 Expression on the Growth and Metastasis of Lewis Lung Carcinoma. Oncol. Lett. 2019, 18, 1548–1556. [Google Scholar] [CrossRef]
- Li, J.; Qu, X.; Tian, J.; Zhang, J.-T.; Cheng, J.-X. Cholesterol Esterification Inhibition and Gemcitabine Synergistically Suppress Pancreatic Ductal Adenocarcinoma Proliferation. PLoS ONE 2018, 13, e0193318. [Google Scholar] [CrossRef] [Green Version]
- Delsing, D.J.; Offerman, E.H.; van Duyvenvoorde, W.; van Der Boom, H.; de Wit, E.C.; Gijbels, M.J.; van Der Laarse, A.; Jukema, J.W.; Havekes, L.M.; Princen, H.M. Acyl-Coa:Cholesterol Acyltransferase Inhibitor Avasimibe Reduces Atherosclerosis in Addition to Its Cholesterol-Lowering Effect in Apoe*3-Leiden Mice. Circulation 2001, 103, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Sahi, J.; Milad, M.A.; Zheng, X.; Rose, K.A.; Wang, H.; Stilgenbauer, L.; Gilbert, D.; Jolley, S.; Stern, R.H.; Lecluyse, E.L. Avasimibe Induces Cyp3a4 and Multiple Drug Resistance Protein 1 Gene Expression through Activation of the Pregnane X Receptor. J. Pharmacol. Exp. Ther. 2003, 306, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).