Expression of the Nonclassical MHC Class I, Saha-UD in the Transmissible Cancer Devil Facial Tumour Disease (DFTD)
Abstract
1. Introduction
2. Results
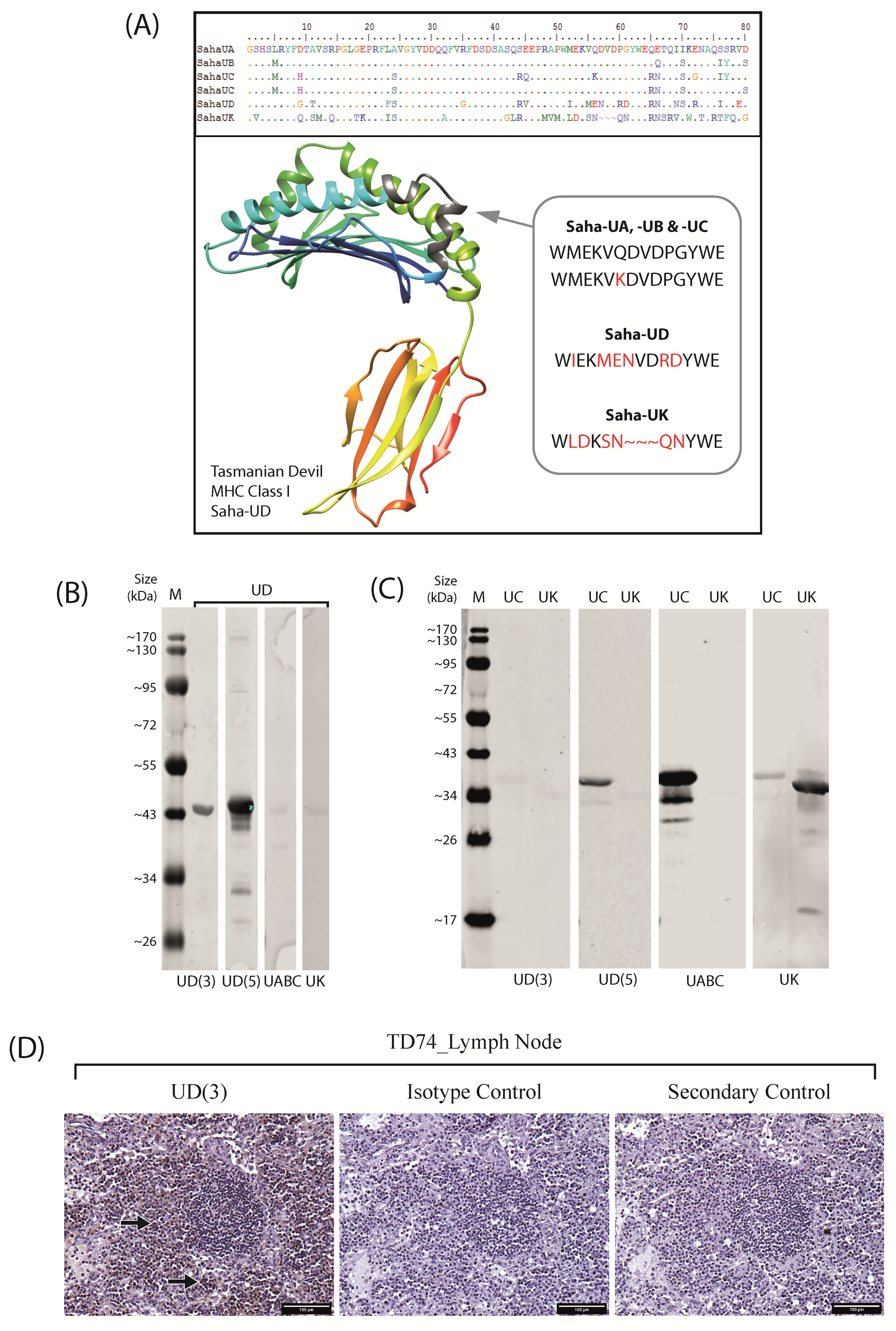
2.1. Anti-Saha-UD Antibody, α-14-37-3, Specifically Binds Recombinant Nonclassical Saha-UD Protein
2.2. Nonclassical Saha-UD Is Expressed in Primary DFTD Tumours, but Expression Varies and Tumours Show Intratumour Heterogeneity
2.3. Distinct Patterns of Staining for Nonclassical Saha-UD versus Classical (Saha-UA, -UB, & -UC)
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Sequence Analysis
4.3. Anti-Saha-UD Antibody Generation
4.4. Recombinant MHC Class I Proteins
4.5. SDS-PAGE and Western Blot
4.6. Immunohistochemistry (IHC)
4.7. RT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| β2m | β2-microglobulin |
| CTVT | Canine Transmissible Venereal Tumour |
| DFTD | Devil Facial Tumour Disease 1 |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| HLA | Human Leukocyte Antigen |
| IFN-γ | Interferon-γ |
| IHC | Immunohistochemistry |
| NK Cell | Natural Killer Cell |
| MHC-I | Major Histocompatibility Complex Class I |
| PBS | Phosphate Buffered Saline |
| PCR | Polymerase Chain Reaction |
| PRC2 | Polycomb Repressive Complex 2 |
| TAP | Transporter Associated with Antigen Processing |
References
- Pearse, A.; Swift, K.A.D. Transmission of devil facial-tumour disease. Nature 2006, 439, 549. [Google Scholar] [CrossRef] [PubMed]
- Loh, R.; Bergfeld, J.; Hayes, D.; O’Hara, A.; Pyecroft, S.; Raidal, S.; Sharpe, R. The Pathology of Devil Facial Tumor Disease (DFTD) in Tasmanian Devils (Sarcophilus harrisii). Vet. Pathol. 2006, 43, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Hamede, R.K.; McCallum, H.; Jones, M. Seasonal, demographic and density-related patterns of contact between Tasmanian devils (Sarcophilus harrisii): Implications for transmission of devil facial tumour disease. Austral. Ecol. 2008, 33, 614–622. [Google Scholar] [CrossRef]
- Hamede, R.K.; McCallum, H.; Jones, M. Biting injuries and transmission of Tasmanian devil facial tumour disease. J. Anim. Ecol. 2013, 82, 182–190. [Google Scholar] [CrossRef]
- Hawkins, C.; Baars, C.; Hesterman, H.; Hocking, G.; Jones, M.; Lazenby, B.; Mann, D.; Mooney, N.; Pemberton, D.; Pyecroft, S.; et al. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol. Conserv. 2006, 131, 307–324. [Google Scholar] [CrossRef]
- Lazenby, B.T.; Tobler, M.W.; Brown, W.E.; Hawkins, C.E.; Hocking, G.J.; Hume, F.; Huxtable, S.; Iles, P.; Jones, M.; Lawrence, C.; et al. Density trends and demographic signals uncover the long-term impact of transmissible cancer in Tasmanian devils. J. Appl. Ecol. 2018, 55, 1368–1379. [Google Scholar] [CrossRef]
- Hawkins, C.E.; McCallum, H.; Mooney, N.; Jones, M.; Holdsworth, M. Sarcophilus harrisii. The IUCN Red List of Threatened Species 2008: e.T40540A10331066. 2008. Available online: https://www.iucnredlist.org/species/40540/10331066 (accessed on 30 January 2022).
- Patton, A.H.; Lawrance, M.F.; Margres, M.J.; Kozakiewicz, C.P.; Hamede, R.; Ruiz-Aravena, M.; Hamilton, D.G.; Comte, S.; Ricci, L.E.; Taylor, R.L.; et al. A transmissible cancer shifts from emergence to endemism in Tasmanian devils. Science 2020, 370, eabb9772. [Google Scholar] [CrossRef]
- Epstein, B.; Jones, M.; Hamede, R.; Hendricks, S.; McCallum, H.; Murchison, E.P.; Schönfeld, B.; Wiench, C.; Hohenlohe, P.; Storfer, A. Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Hubert, J.-N.; Zerjal, T.; Hospital, F. Cancer- and behavior-related genes are targeted by selection in the Tasmanian devil (Sarcophilus harrisii). PLoS ONE 2018, 13, e0201838. [Google Scholar] [CrossRef]
- Margres, M.J.; Jones, M.E.; Epstein, B.; Kerlin, D.H.; Comte, S.; Fox, S.; Fraik, A.K.; Hendricks, S.A.; Huxtable, S.; Lachish, S.; et al. Large-effect loci affect survival in Tasmanian devils (Sarcophilus harrisii) infected with a transmissible cancer. Mol. Ecol. 2018, 27, 4189–4199. [Google Scholar] [CrossRef]
- Ruiz-Aravena, M.; Jones, M.E.; Carver, S.; Estay, S.; Espejo, C.; Storfer, A.; Hamede, R.K. Sex bias in ability to cope with cancer: Tasmanian devils and facial tumour disease. Proc. R. Soc. B Biol. Sci. 2018, 285, 20182239. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. T Cells and MHC Proteins. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Siddle, H.V.; Marzec, J.; Cheng, Y.; Jones, M.; Belov, K. MHC gene copy number variation in Tasmanian devils: Implications for the spread of a contagious cancer. Proc. R. Soc. B Biol. Sci. 2010, 277, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Stuart, A.; Morris, K.; Taylor, R.; Siddle, H.; Deakin, J.; Jones, M.; Amemiya, C.T.; Belov, K. Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC. BMC Genom. 2012, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Kreiss, A.; Cheng, Y.; Kimble, F.; Wells, B.; Donovan, S.; Belov, K.; Woods, G.M. Allorecognition in the Tasmanian Devil (Sarcophilus harrisii), an Endangered Marsupial Species with Limited Genetic Diversity. PLoS ONE 2011, 6, e22402. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pye, R.; Hamede, R.; Siddle, H.V.; Caldwell, A.; Knowles, G.W.; Swift, K.; Kreiss, A.; Jones, M.; Lyons, A.; Woods, G.M. Demonstration of immune responses against devil facial tumour disease in wild Tasmanian devils. Biol. Lett. 2016, 12, 20160553. [Google Scholar] [CrossRef] [PubMed]
- Siddle, H.V.; Kreiss, A.; Tovar, C.; Yuen, C.K.; Cheng, Y.; Belov, K.; Swift, K.; Pearse, A.-M.; Hamede, R.; Jones, M.; et al. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 5103–5108. [Google Scholar] [CrossRef]
- Ong, C.E.B.; Patchett, A.L.; Darby, J.M.; Chen, J.; Liu, G.-S.; Lyons, A.B.; Woods, G.M.; Flies, A.S. NLRC5 regulates expression of MHC-I and provides a target for anti-tumor immunity in transmissible cancers. J. Cancer Res. Clin. Oncol. 2021, 147, 1973–1991. [Google Scholar] [CrossRef]
- Burr, M.; Sparbier, C.; Chan, K.L.; Chan, Y.-C.; Kersbergen, A.; Lam, E.Y.; Azidis-Yates, E.; Vassiliadis, D.; Bell, C.; Gilan, O.; et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 2019, 36, 385–401. [Google Scholar] [CrossRef]
- Ljunggren, H.-G.; Kärre, K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 1990, 11, 237–244. [Google Scholar] [CrossRef]
- Brown, G.K.; Kreiss, A.; Lyons, A.B.; Woods, G.M. Natural Killer Cell Mediated Cytotoxic Responses in the Tasmanian Devil. PLoS ONE 2011, 6, e24475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Braud, V.M.; Allan, D.S.; McMichael, A.J. Functions of nonclassical MHC and non-MHC-encoded class I molecules. Curr. Opin. Immunol. 1999, 11, 100–108. [Google Scholar] [CrossRef]
- Janeway, C.A.; Travers, P.; Walport, M.; Shlomchik, M.J. The major histocompatibility complex and its functions. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Allen, R.L.; Hogan, L. Non-Classical MHC Class I Molecules (MHC-Ib). eLS 2013. [Google Scholar] [CrossRef]
- Cheng, Y.; Belov, K. Characterisation of non-classical MHC class I genes in the Tasmanian devil (Sarcophilus harrisii). Immunogenetics 2014, 66, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Papenfuss, A.T.; Feng, Z.-P.; Krasnec, K.; Deakin, J.E.; Baker, M.L.; Miller, R.D. Marsupials and monotremes possess a novel family of MHC class I genes that is lost from the eutherian lineage. BMC Genom. 2015, 16, 535. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, P.J.; Parham, P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu. Rev. Biochem. 1990, 59, 253–288. [Google Scholar] [CrossRef]
- Gastaldello, A.; Ramarathinam, S.H.; Bailey, A.; Owen, R.; Turner, S.; Kontouli, N.; Elliott, T.; Skipp, P.; Purcell, A.W.; Siddle, H.V. The immunopeptidomes of two transmissible cancers and their host have a common, dominant peptide motif. Immunology 2021, 163, 169–184. [Google Scholar] [CrossRef]
- Caldwell, A.; Siddle, H.V. The role of MHC genes in contagious cancer: The story of Tasmanian devils. Immunogenetics 2017, 69, 537–545. [Google Scholar] [CrossRef]
- Caldwell, A.; Coleby, R.; Tovar, C.; Stammnitz, M.R.; Kwon, Y.M.; Owen, R.S.; Tringides, M.; Murchison, E.P.; Skjødt, K.; Thomas, G.J.; et al. The newly-arisen Devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer. eLife 2018, 7, e35314. [Google Scholar] [CrossRef]
- Pye, R.; Pemberton, D.; Tovar, C.; Tubio, J.; Dun, K.; Fox, S.; Darby, J.; Hayes, D.; Knowles, G.W.; Kreiss, A.; et al. A second transmissible cancer in Tasmanian devils. Proc. Natl. Acad. Sci. USA 2016, 113, 374–379. [Google Scholar] [CrossRef]
- Kwon, Y.M.; Gori, K.; Park, N.; Potts, N.; Swift, K.; Wang, J.; Stammnitz, M.R.; Cannell, N.; Baez-Ortega, A.; Comte, S.; et al. Evolution and lineage dynamics of a transmissible cancer in Tasmanian devils. PLoS Biol. 2020, 18, e3000926. [Google Scholar] [CrossRef] [PubMed]
- Kreiss, A.; Brown, G.; Tovar, C.; Lyons, A.; Woods, G. Evidence for induction of humoral and cytotoxic immune responses against devil facial tumor disease cells in Tasmanian devils (Sarcophilus harrisii) immunized with killed cell preparations. Vaccine 2015, 33, 3016–3025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patchett, A.L.; Tovar, C.; Blackburn, N.B.; Woods, G.M.; Lyons, A.B. Mesenchymal plasticity of devil facial tumour cells during in vivo vaccine and immunotherapy trials. Immunol. Cell Biol. 2021, 99, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Murgia, C.; Pritchard, J.K.; Kim, S.Y.; Fassati, A.; Weiss, R.A. Clonal Origin and Evolution of a Transmissible Cancer. Cell 2006, 126, 477–487. [Google Scholar] [CrossRef]
- Belov, K. The role of the Major Histocompatibility Complex in the spread of contagious cancers. Mamm. Genome 2011, 22, 83–90. [Google Scholar] [CrossRef]
- Tovar, C.; Pye, R.; Kreiss, A.; Cheng, Y.; Brown, G.K.; Darby, J.; Malley, R.C.; Siddle, H.V.T.; Skjødt, K.; Kaufman, J.; et al. Regression of devil facial tumour disease following immunotherapy in immunised Tasmanian devils. Sci. Rep. 2017, 7, srep43827. [Google Scholar] [CrossRef]
- Djurisic, S.; Hviid, T.V.F. HLA Class Ib Molecules and Immune Cells in Pregnancy and Preeclampsia. Front. Immunol. 2014, 5, 652. [Google Scholar] [CrossRef]
- Tersigni, C.; Meli, F.; Neri, C.; Iacoangeli, A.; Franco, R.; Lanzone, A.; Scambia, G.; Di Simone, N. Role of Human Leukocyte Antigens at the Feto-Maternal Interface in Normal and Pathological Pregnancy: An Update. Int. J. Mol. Sci. 2020, 21, 4756. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Murchison, E.P.; Tovar, C.; Hsu, A.; Bender, H.S.; Kheradpour, P.; Rebbeck, C.A.; Obendorf, D.; Conlan, C.; Bahlo, M.; Blizzard, C.A.; et al. The Tasmanian Devil Transcriptome Reveals Schwann Cell Origins of a Clonally Transmissible Cancer. Science 2010, 327, 84–87. [Google Scholar] [CrossRef]
- Tovar, C.; Obendorf, D.; Murchison, E.P.; Papenfuss, A.T.; Kreiss, A.; Woods, G.M. Tumor-Specific Diagnostic Marker for Transmissible Facial Tumors of Tasmanian Devils: Immunohistochemistry Studies. Vet. Pathol. 2011, 48, 1195–1203. [Google Scholar] [CrossRef] [PubMed]



| MHC Class I | Classical (SahaI-01) | Saha-UD (SahaI*12) | Saha-UK | Saha-UM |
|---|---|---|---|---|
| Classical (SahaI-01) | 78.2% | 60.6% | 52.1% | |
| Saha-UD (SahaI*12) | 78.2% | 58.1% | 52.4% | |
| Saha-UK | 60.6% | 58.1% | 43.1% | |
| Saha-UM | 52.1% | 52.4% | 43.1% |
| Gene Amplified | Primer Names | Primer Sequence (5′ -> 3′) | Product Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| Nonclassical MHC Class I Saha-UD (exon 2–4) | Saha138 | GCCATATGGGCTCTCACTCCTTGAGGTATTTCGGCACCAC | 1044 | 63 |
| Saha48 | ATGCAAGCTTGGCTTTGGCTGTCAGAGAGACATCTGACC | |||
| RPL13A | Saha120 | ACAAGACCAAGCGAGGCCAGG | 300 | 60 |
| Saha121 | GCCTGGTATTTCCAGCCAACCTCA | |||
| Nonclassical MHC Class I Saha-UD | Dev Men UD (F) | ATGGAGAATGTGGACCGGGAC | 275 | 60 |
| Dev Men UD (R) | TGAGTTCACTGCCTCATTCACT |
| PCR Reagents | RCR Reaction Conditions | ||||
|---|---|---|---|---|---|
| Reagent | Final Concentration | Cycle Element | Temperature (°C) | Time (s) | Number of Cycles |
| cDNA | 500 ng | Initial denaturation | 98 | 30 | 1 |
| DNA polymerase (Thermo Fisher Phusion) | 0.5 U | Denaturation | 98 | 10 | 30 |
| Primers | 0.6 µM | Annealing | Temperatures for primers in Table 2 | 30 | |
| dNTPs | 200 µM | Elongation | 72 | 30 | |
| Phusion High Fidelity buffer | 1X | Final Elongation | 72 | 300 | 1 |
| ddH2O | To total volume of 25 µl | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussey, K.; Caldwell, A.; Kreiss, A.; Skjødt, K.; Gastaldello, A.; Pye, R.; Hamede, R.; Woods, G.M.; Siddle, H.V. Expression of the Nonclassical MHC Class I, Saha-UD in the Transmissible Cancer Devil Facial Tumour Disease (DFTD). Pathogens 2022, 11, 351. https://doi.org/10.3390/pathogens11030351
Hussey K, Caldwell A, Kreiss A, Skjødt K, Gastaldello A, Pye R, Hamede R, Woods GM, Siddle HV. Expression of the Nonclassical MHC Class I, Saha-UD in the Transmissible Cancer Devil Facial Tumour Disease (DFTD). Pathogens. 2022; 11(3):351. https://doi.org/10.3390/pathogens11030351
Chicago/Turabian StyleHussey, Kathryn, Alison Caldwell, Alexandre Kreiss, Karsten Skjødt, Annalisa Gastaldello, Ruth Pye, Rodrigo Hamede, Gregory M. Woods, and Hannah V. Siddle. 2022. "Expression of the Nonclassical MHC Class I, Saha-UD in the Transmissible Cancer Devil Facial Tumour Disease (DFTD)" Pathogens 11, no. 3: 351. https://doi.org/10.3390/pathogens11030351
APA StyleHussey, K., Caldwell, A., Kreiss, A., Skjødt, K., Gastaldello, A., Pye, R., Hamede, R., Woods, G. M., & Siddle, H. V. (2022). Expression of the Nonclassical MHC Class I, Saha-UD in the Transmissible Cancer Devil Facial Tumour Disease (DFTD). Pathogens, 11(3), 351. https://doi.org/10.3390/pathogens11030351






