Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies
Abstract
1. Introduction
2. Results
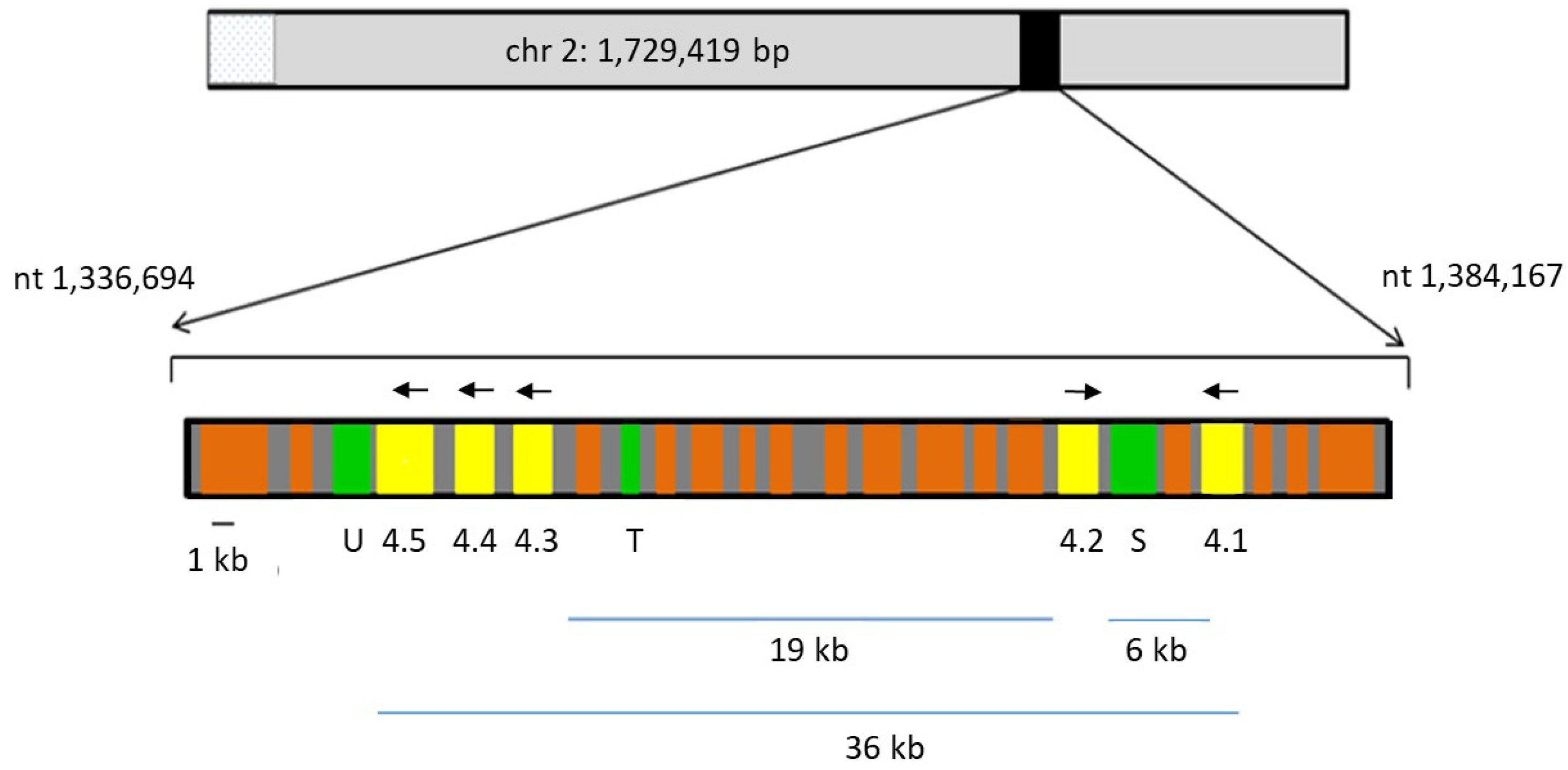
2.1. Genomic Location of rom4 Paralogs
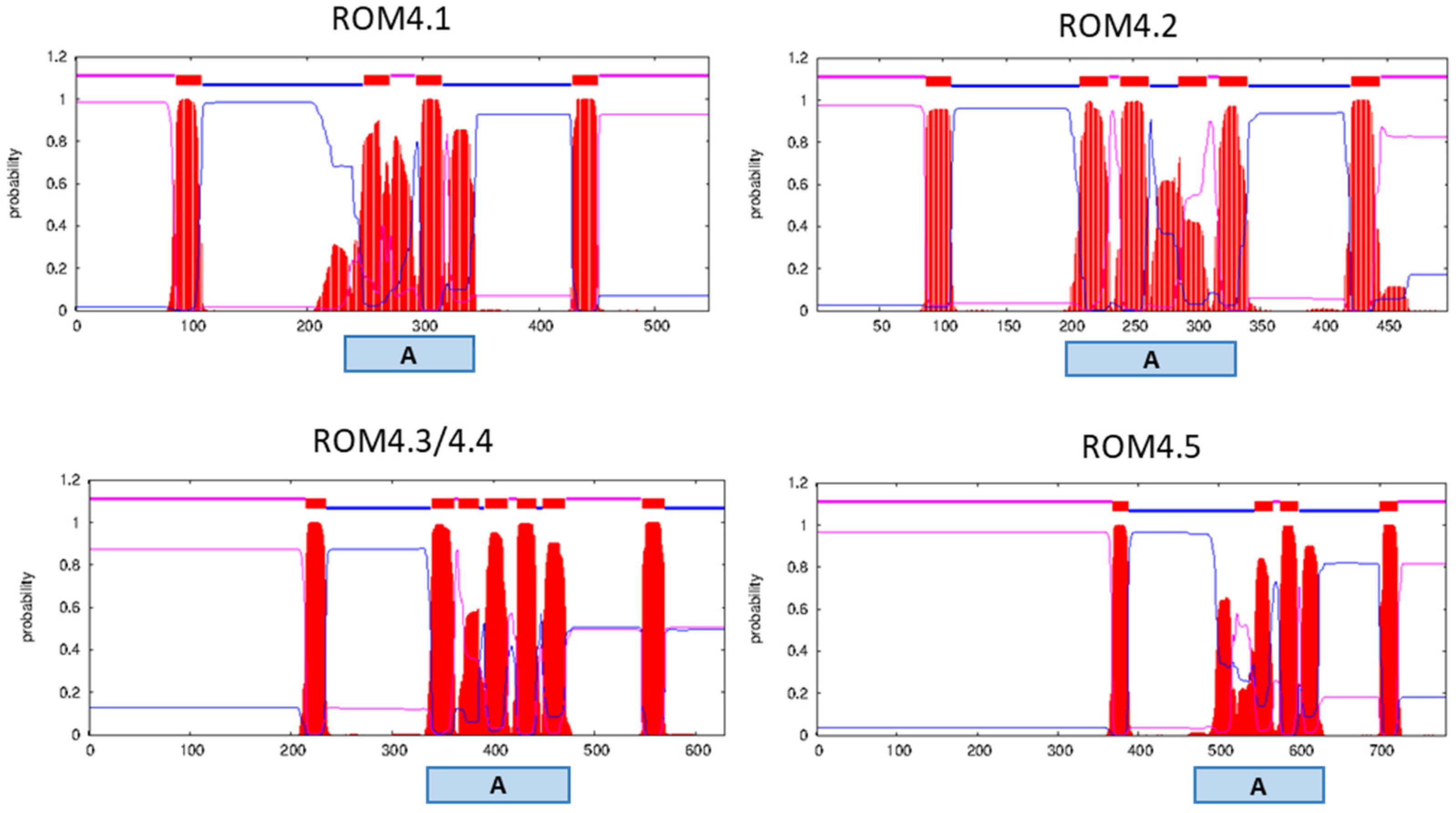
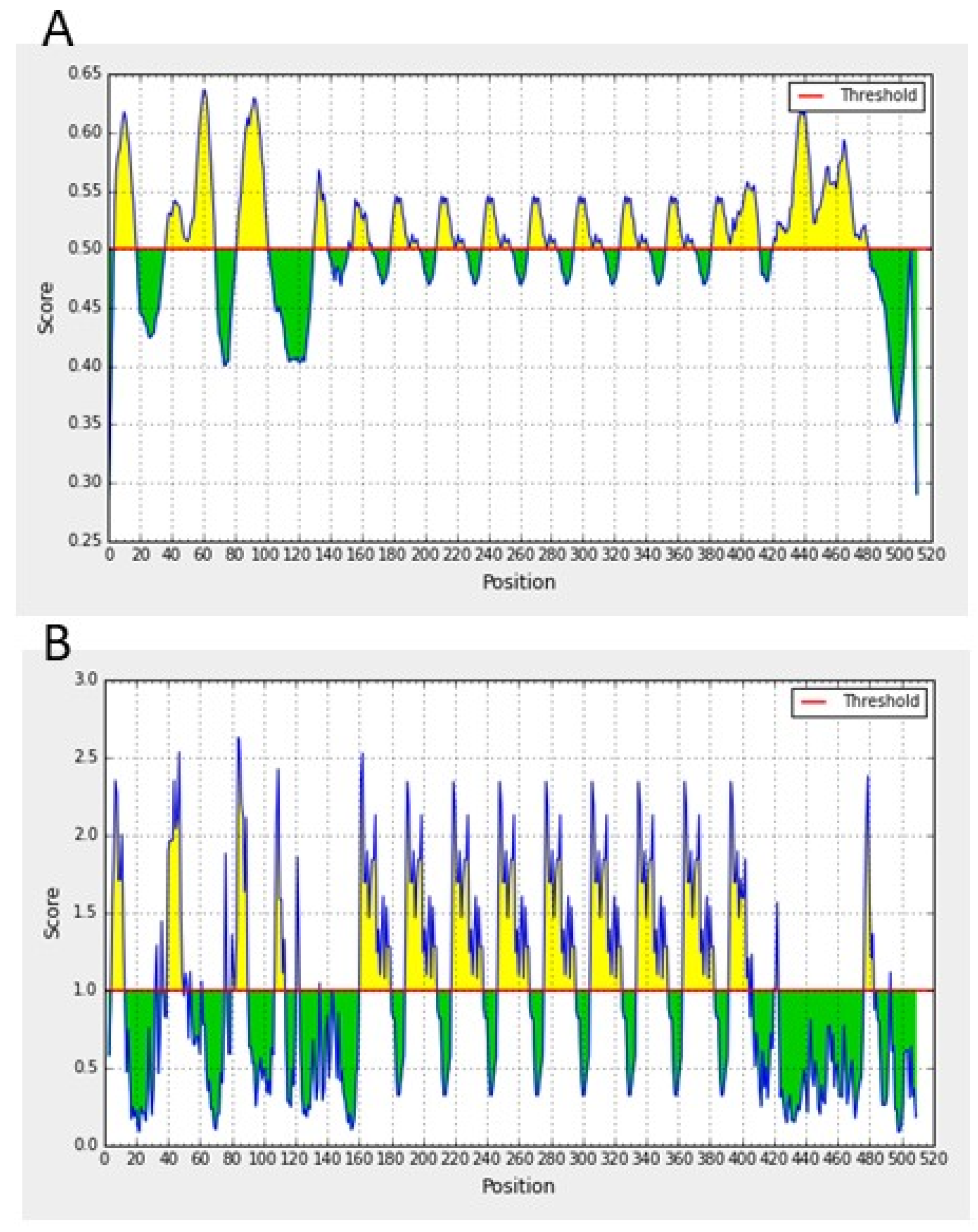
2.2. In Silico Characterization of ROM4 Proteins
2.3. Synteny Analysis of B. bovis rom4 Paralogs
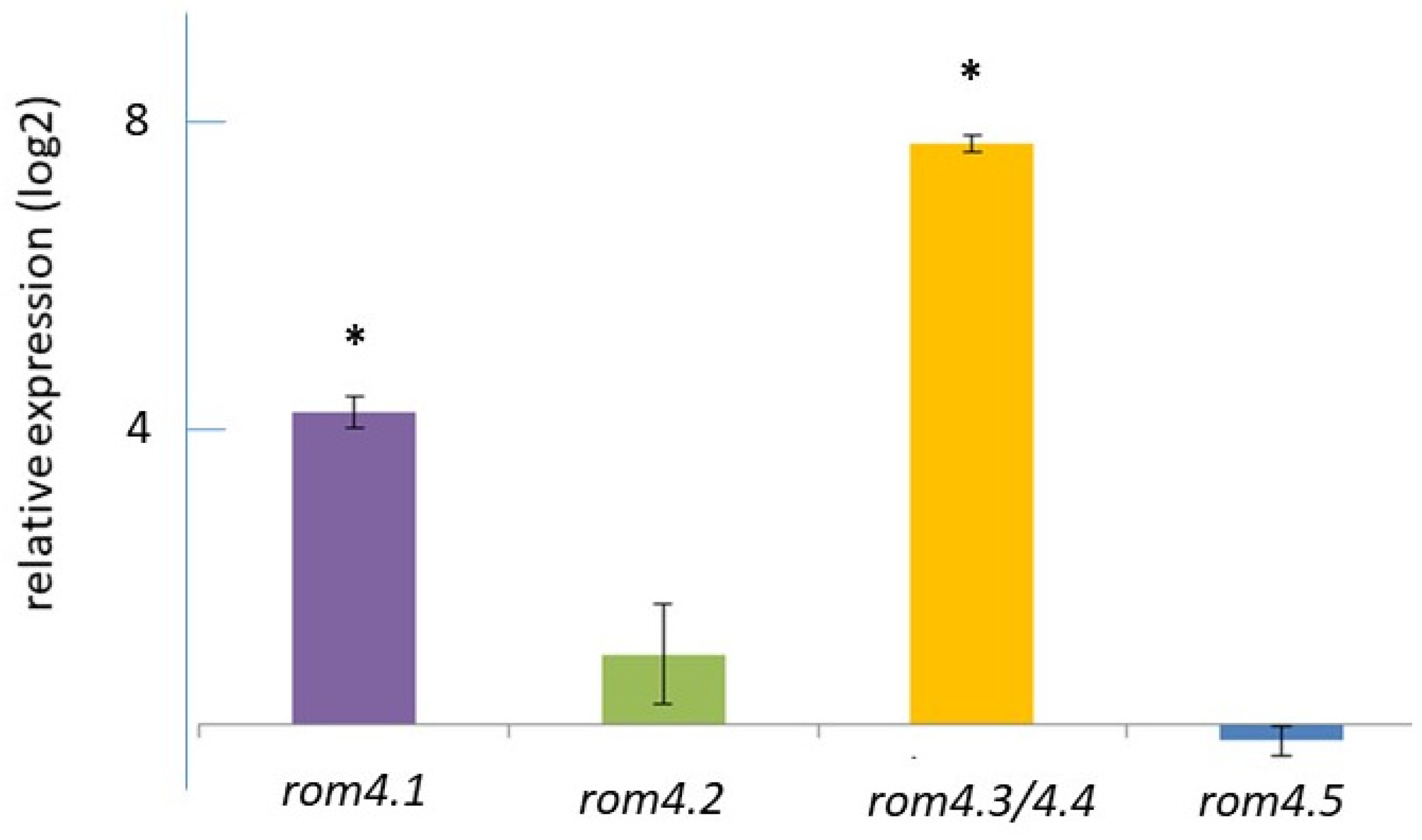
2.4. Expression of Two rom4 Genes Is Significantly Increased in Xanthurenic Acid-Induced Xexual Stages of B. bovis Compared with Non-Induced Merozoite Suspensions
2.5. Sequence Conservation of ROM4 Paralogs among Distinct B. bovis Geographic Isolates
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis
4.2. Production of cDNA of In Vitro Cultured AS and SS
4.3. Transcriptional Analysis
4.4. DNA Samples
4.5. Sequence Analysis of rom4 Alleles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Florin-Christensen, M.; Suarez, C.E.; Rodriguez, A.E.; Flores, D.A.; Schnittger, L. Vaccines against bovine babesiosis: Where we are now and possible roads ahead. Parasitology 2014, 141, 1563–1592. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Ganzinelli, S.; Bhoora, R.; Omondi, D.; Nijhof, A.M.; Florin-Christensen, M. The Piroplasmida Babesia, Cytauxzoon, and Theileria in farm and companion animals: Species compilation, molecular phylogeny, and evolutionary insights. Parasitol. Res. 2022, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Jalovecka, M.; Hajdusek, O.; Sojka, D.; Kopacek, P.; Malandrin, L. The complexity of piroplasms life cycles. Front. Cell. Infect. Microbiol. 2018, 8, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Bock, R.; Jackson, L.; de Vos, A.; Jorgensen, W. Babesiosis of cattle. Parasitology 2004, 129, S247–S269. [Google Scholar] [CrossRef] [PubMed]
- Ganzinelli, S.; Rodriguez, A.E.; Schnittger, L.; Florin-Christensen, M. Babesia in domestic ruminants. In Parasitic Protozoa of Farm Animals and Pets; Florin-Christensen, M., Schnittger, L., Eds.; Springer International Publishing: New York, NY, USA, 2018; pp. 215–240. [Google Scholar]
- Trueman, K.F.; McLennan, M.W. Bovine abortion due to prenatal Babesia bovis infection. Aust. Vet. J. 1987, 64, 63. [Google Scholar] [CrossRef] [PubMed]
- Florin-Christensen, M.; Schnittger, L. Piroplasmids and ticks: A long-lasting intimate relationship. Front. Biosci. 2009, 14, 3064–3073. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.E.; Alzan, H.F.; Silva, M.G.; Rathinasamy, V.; Poole, W.A.; Cooke, B.M. Unravelling the cellular and molecular pathogenesis of bovine babesiosis: Is the sky the limit? Int. J. Parasitol. 2019, 49, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Klemba, M.; Goldberg, D.E. Biological roles of proteases in parasitic protozoa. Annu. Rev. Biochem. 2002, 71, 275–305. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M. Proteolysis within the membrane: Rhomboids revealed. Nat. Rev. Mol. Cell Biol. 2004, 5, 188–197. [Google Scholar] [CrossRef]
- Dowse, T.J.; Soldati, D. Rhomboid-like Proteins in Apicomplexa: Phylogeny and Nomenclature. Trends Parasitol. 2005, 21, 254–258. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, J.; Meireles, P.; Prudêncio, M.; Engelmann, S.; Annoura, T.; Sajid, M.; Chevalley-Maurel, S.; Ramesar, J.; Nahar, C.; Avramut, C.M.C.; et al. Loss-of-function analyses defines vital and redundarnt functions of the Plasmodium rhomboid protease family. Mol. Microbiol. 2013, 88, 318–338. [Google Scholar] [CrossRef] [PubMed]
- Dogga, S.K.; Soldati-Favre, D. Biology of Rhomboid Proteases in Infectious Diseases. Semin. Cell Dev. Biol. 2016, 60, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Düsterhöft, S.; Künzel, U.; Freeman, M. Rhomboid Proteases in Human Disease: Mechanisms and Future Prospects. Biochim. Biophys. Acta 2017, 1864, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Gallenti, R.; Poklepovich, T.; Florin-Christensen, M.; Schnittger, L. The repertoire of serine rhomboid proteases of piroplasmids of importance to animal and human health. Int. J. Parasitol. 2021, 51, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Ueti, M.W.; Johnson, W.C.; Kappmeyer, L.S.; Herndon, D.R.; Mousel, M.R.; Reif, K.E.; Taus, N.S.; Ifeonu, O.O.; Silva, J.C.; Suarez, C.E.; et al. Transcriptome dataset of Babesia bovis life stages within vertebrate and invertebrate hosts. Data Brief 2020, 33, 106533. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Rogozin, I.B.; Davidovic, L.; Letellier, M.C.; Pellegrini, L. The rhomboids: A nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003, 4, R19. [Google Scholar] [CrossRef]
- Urban, S.; Freeman, M. Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev. 2002, 12, 512–518. [Google Scholar] [CrossRef]
- Santos, J.M.; Graindorge, A.; Soldati-Favre, D. New Insights into Parasite Rhomboid Proteases. Mol. Biochem. Parasitol. 2012, 182, 27–36. [Google Scholar] [CrossRef]
- Gaffar, F.R.; Yatsuda, A.P.; Franssen, F.F.J.; De Vries, E. Erythrocyte invasion by Babesia bovis merozoites is inhibited by polyclonal antisera directed against peptides derived from a homologue of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 2004, 72, 2947–2955. [Google Scholar] [CrossRef]
- Gaffar, F.R.; Yatsuda, A.P.; Franssen, F.F.J.; De Vries, E.A. Babesia bovis merozoite protein with a domain architecture highly similar to the thrombospondin-related anonymous protein (TRAP) present in Plasmodium sporozoites. Mol. Biochem. Parasitol. 2004, 136, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Billker, O.; Lindo, V.S.; Panico, M.; Etienne, A.; Paxton, T.; Dell, A.; Rogers, M.C.; E Sinden, R.; Morris, H.R. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature 1998, 392, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.E.; Wirtz, R.A.; Barr, J.R.; Woolfitt, A.; Rosenberg, R. Xanthurenic acid induces gametogenesis in Plasmodium, the malaria parasite. J. Biol. Chem. 1998, 273, 12003–12005. [Google Scholar] [CrossRef]
- Hussein, H.E.; Bastos, R.G.; Schneider, D.A.; Johnson, W.C.; Adham, F.K.; Davis, W.C.; Laughery, J.M.; Herndon, D.R.; Alzan, H.F.; Ueti, M.W.; et al. The Babesia bovis hap2 gene is not required for blood stage replication, but expressed upon in vitro sexual stage induction. PLoS Negl. Trop. Dis. 2017, 11, e0005965. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.E.; Johnson, W.C.; Taus, N.S.; Capelli-Peixoto, J.; Suarez, C.E.; Mousel, M.R.; Ueti, M.W. Differential expression of calcium-dependent protein kinase 4, tubulin tyrosine ligase, and methyltransferase by xanthurenic acid-induced Babesia bovis sexual stages. Parasites Vectors 2021, 14, 395. [Google Scholar] [CrossRef] [PubMed]
- Mosqueda, J.; Falcon, A.; Alvarez, A.J.; Ramos, A.J.; Oropeza-Hernandez, L.F.; Figueroa, J.V. Babesia bigemina sexual stages are induced in vitro and are specifically recognized by antibodies in the midgut of infected Boophilus microplus ticks. Int. J. Parasitol. 2004, 34, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Dang-Trinh, M.A.; Higuchi, L.; Mosqueda, J.; Hakimi, H.; Asada, M.; Yamagishi, J.; Umemiya-Shirafuji, R.; Kawazu, S.I. Initiated Babesia ovata Sexual Stages under In Vitro Conditions Were Recognized by Anti-CCp2 Antibodies, Showing Changes in the DNA Content by Imaging Flow Cytometry. Pathogens 2019, 8, 104. [Google Scholar] [CrossRef]
- Schnittger, L.; Katzer, F.; Biermann, R.; Shayan, P.; Boguslawski, K.; McKellar, S.; Beyer, D.; Shiels, B.R.; Ahmed, J.S. Characterization of a polymorphic Theileria annulata surface protein (TaSP) closely related to PIM of Theileria parva: Implications for use in diagnostic tests and subunit vaccines. Mol. Biochem. Parasitol. 2002, 120, 247–256. [Google Scholar] [CrossRef]
- Suarez, C.E.; Laughery, J.M.; Bastos, R.G.; Johnson, W.C.; Norimine, J.; Asenzo, G.; Brown, W.C.; Florin-Christensen, M.; Goff, W.L. A novel neutralization sensitive and subdominant RAP-1-related antigen (RRA) is expressed by Babesia bovis merozoites. Parasitology 2011, 138, 809–818. [Google Scholar] [CrossRef]
- Mesplet, M.; Palmer, G.H.; Pedroni, M.J.; Echaide, I.; Florin-Christensen, M.; Schnittger, L.; Lau, A.O. Genome-wide analysis of peptidase content and expression in a virulent and attenuated Babesia bovis strain pair. Mol. Biochem. Parasitol. 2011, 179, 111–113. [Google Scholar] [CrossRef]
- Lysyk, L.; Brassard, R.; Touret, N.; Lemieux, M.J. PARL Protease: A Glimpse at Intramembrane Proteolysis in the Inner Mitochondrial Membrane. J. Mol. Biol. 2020, 432, 5052–5062. [Google Scholar] [CrossRef] [PubMed]
- Nejatfard, A.; Wauer, N.; Bhaduri, S.; Conn, A.; Gourkanti, S.; Singh, N.; Kuo, T.; Kandel, R.; Amaro, R.E.; Neal, S.E. Derlin rhomboid pseudoproteases employ substrate engagement and lipid distortion to enable the retrotranslocation of ERAD membrane substrates. Cell Rep. 2021, 37, 109840. [Google Scholar] [CrossRef] [PubMed]
- Florin-Christensen, M.; Schnittger, L.; Bastos, R.G.; Rathinasamy, V.A.; Cooke, B.M.; Alzan, H.F.; Suarez, C.E. Pursuing effective vaccines against cattle diseases caused by apicomplexan protozoa. CAB Rev. 2021, 16, 24. [Google Scholar] [CrossRef]
- Bastos, R.G.; Suarez, C.E.; Laughery, J.M.; Johnson, W.C.; Ueti, M.W.; Knowles, D.P. Differential expression of three members of the multidomain adhesion CCp family in Babesia bigemina, Babesia bovis and Theileria equi. PLoS ONE 2013, 8, e67765. [Google Scholar] [CrossRef] [PubMed]
- Alzan, H.F.; Bastos, R.G.; Ueti, M.W.; Laughery, J.M.; Rathinasamy, V.A.; Cooke, B.M.; Suarez, C.E. Assessment of Babesia bovis 6cys A and 6cys B as components of transmission blocking vaccines for babesiosis. Parasites Vectors 2021, 14, 210. [Google Scholar] [CrossRef] [PubMed]
- Alzan, H.F.; Cooke, B.M.; Suarez, C.E. Transgenic Babesia bovis lacking 6-Cys sexual-stage genes as the foundation for non-transmissible live vaccines against bovine babesiosis. Ticks Tick Borne Dis. 2019, 10, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Alzan, H.F.; Lau, A.O.; Knowles, D.P.; Herndon, D.R.; Ueti, M.W.; Scoles, G.A.; Kappmeyer, L.S.; Suarez, C.E. Expression of 6-Cys gene superfamily defines Babesia bovis sexual stage development within Rhipicephalus microplus. PLoS ONE 2016, 11, e0163791. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, J.; Gong, P.; Zhang, X. Efficacy of Eimeria tenella rhomboid-like protein as a subunit vaccine in protective immunity against homologous challenge. Parasitol. Res. 2012, 110, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.Z.; Xu, Y.; Wang, M.; Petersen, E.; Chen, J.; Huang, S.Y.; Zhu, X.Q. Protective efficacy of two novel DNA vaccines expressing Toxoplasma gondii rhomboid 4 and rhomboid 5 proteins against acute and chronic toxoplasmosis in mice. Expert Rev. Vaccines 2015, 14, 1289–1297. [Google Scholar] [CrossRef]
- Han, Y.; Zhou, A.; Lu, G.; Zhao, G.; Wang, L.; Guo, J.; Song, P.; Zhou, J.; Zhou, H.; Cong, H.; et al. Protection via a ROM4 DNA vaccine and peptide against Toxoplasma gondii in BALB/c mice. BMC Infect. Dis. 2017, 17, 59–68. [Google Scholar] [CrossRef]
- Urban, S. Making the cut: Central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 2009, 7, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Strisovsky, K. Rhomboid protease inhibitors: Emerging tools and future therapeutics. Semin. Cell Dev. Biol. 2016, 60, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Tichá, A.; Stanchev, S.; Vinothkumar, K.R.; Mikles, D.C.; Pachl, P.; Began, J.; Škerle, J.; Švehlová, K.; Nguyen, M.T.N.; Verhelst, S.H.L.; et al. General and modular strategy for designing potent, selective, and pharmacologically compliant inhibitors of rhomboid proteases. Cell Chem. Biol. 2017, 24, 1523–1536.e4. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Baker, R.P.; Cho, S.; Stanchev, S.; Strisovsky, K.; Urban, S. Designed parasite-selective rhomboid inhibitors block invasion and clear blood-stage malaria. Cell Chem. Biol. 2020, 27, 1410–1424.e6. [Google Scholar] [CrossRef] [PubMed]
- Brayton, K.A.; Lau, A.O.; Herndon, D.R.; Hannick, L.; Kappmeyer, L.S.; Berens, S.J.; Bidwell, S.L.; Brown, W.C.; Crabtree, J.; Fadrosh, D.; et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007, 3, 1401–1413. [Google Scholar] [CrossRef]
- Bustin, S.A.; Beaulieu, J.F.; Huggett, J.; Jaggi, R.; Kibenge, F.S.; Olsvik, P.A.; Penning, L.C.; Toegel, S. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 2010, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Di Rienzo, J.A. fgStatistics. In Statistical Software for the Analysis of Experiments of Functional Genomics; RDNDA: Buenos Aires, Argentina, 2009; p. 756587. Available online: https://sites.google.com/site/fgstatistics/ (accessed on 1 September 2021).
- Hines, S.A.; Palmer, G.H.; Jasmer, D.P.; McGuire, T.C.; McElwain, T.F. Neutralization-sensitive merozoite surface antigens of Babesia bovis encoded by members of a polymorphic gene family. Mol. Biochem. Parasitol. 1992, 55, 85–94. [Google Scholar] [CrossRef]
- Rodriguez, S.D.; Buening, G.M.; Green, T.J.; Carson, C.A. Cloning of Babesia bovis by in vitro cultivation. Infect. Immun. 1983, 42, 15–18. [Google Scholar] [CrossRef]
- Anziani, O.S.; Guglielmone, A.A.; Abdala, A.A.; Aguirre, D.H.; Mangold, A.J. Protección conferida por Babesia bovis vacunal en novillos Holando Argentino. Rev. Med. Vet. 1993, 74, 47–49. [Google Scholar]
- Flores, D.A.; Minichiello, Y.; Araujo, F.R.; Shkap, V.; Benítez, D.; Echaide, I.; Rolls, P.; Mosqueda, J.; Pacheco, G.M.; Petterson, M.; et al. Evidence for extensive genetic diversity and substructuring of the Babesia bovis metapopulation. Transbound. Emerg. Dis. 2013, 60 (Suppl. 2), 131–136. [Google Scholar] [CrossRef]
- Echaide, I.E.; de Echaide, S.T.; Mangold, A.J.; Guglielmone, A.A. Live and soluble antigens from in vitro culture to vaccinate cattle against Babesia bovis. In Proceedings of the IX International Veterinary Hemoparasite Disease Conference, Merida, Mexico, 6–9 October 1993; p. 13. [Google Scholar]
- Ramos, C.A.N.; Araujo, F.R.; Alves, L.C.; Fernando de Souza, I.I.; Guedes, D.S., Jr.; Soares, C.O. Genetic conservation of potentially immunogenic proteins among Brazilian isolates of Babesia bovis. Vet. Parasitol. 2012, 187, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Mazuz, M.L.; Molad, T.; Fish, L.; Leibovitz, B.; Wolkomirsky, R.; Fleiderovitz, L.; Shkap, V. 2012: Genetic diversity of Babesia bovis in virulent and attenuated strains. Parasitology 2012, 139, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Bock, R.E.; Blight, G.W.; Kingston, T.G.; de Vos, A.J. A survey of cattle producers in the Boophilus microplus endemic area of Queensland to determine attitudes to the control of and vaccination against tick fever. Aust. Vet. J. 1995, 72, 88–92. [Google Scholar] [CrossRef] [PubMed]



| Proposed Designation | Protein ID | Size (aa) | MEROPS ID | Active Site (aa) | Gene Locus |
|---|---|---|---|---|---|
| ROM4.1 | EDO06560.2 * | 547 | MER1081995 | 238–341 | BBOV_II006100 |
| ROM4.2 | EDO06557.2 * | 498 | MER1079457 | 193–330 | BBOV_II006070 |
| ROM4.3 | XP_001610113 | 629 | MER0112671 | 308–486 | BBOV_II005950 |
| ROM4.4 | XP_001610112 | 641 | BBOV_II005940 | ||
| ROM4.5 | XP_001610111 | 783 | MER1075434 | 468–621 | BBOV_II005930 |
| ROM4.1 | ROM4.2 | ROM4.3 | ROM4.4 | |
|---|---|---|---|---|
| ROM4.2 | 26.9/37.3 | - | - | - |
| ROM4.3 | 25.6/36.2 | 33.1/45.4 | - | - |
| ROM4.4 | 25.8/36.4 | 33.3/45.6 | 100/100 | - |
| ROM4.5 | 25.9/35.6 | 33.1/44.8 | 73.9/78.7 | 72.5/77.2 |
| Phylogenetic Placement | Piroplasmid Species | B. bovis rom4 Gene Paralogs | ||||
|---|---|---|---|---|---|---|
| rom4.1 | rom4.2 | rom4.3 | rom4.4 | rom4.5 | ||
| Babesia s.s. (Clade VI) | B. bigemina | BBBOND_0104040 | - | - | BBBOND_0104210 | |
| B. ovata | BOVATA_022110 | - | - | BOVATA_021930 | ||
| B. divergens | Bdiv_004020c | Bdiv_003990 | - | Bdiv_003860c | ||
| Babesia sp. Xinjiang | BXIN_2807 | BXIN_2802 | - | BXIN_2789 | ||
| Babesia s.l. (Clade I) | B. microti | BMR1_02g02777 | - | - | - | - |
| Theileria s.s. (Clade V) | T. annulata | TA13905 | - | - | - | - |
| T. orientalis | TOT_020000558 | - | - | - | - | |
| T. parva | TpMuguga_02g02170 | - | - | - | - | |
| Theileria s.l. (Clade IV) | T. equi | BEWA_023690 | - | - | - | - |
| Cytauxzoon (Clade IIIb) | C. felis | CF003707 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallenti, R.; Hussein, H.E.; Alzan, H.F.; Suarez, C.E.; Ueti, M.; Asurmendi, S.; Benitez, D.; Araujo, F.R.; Rolls, P.; Sibeko-Matjila, K.; et al. Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies. Pathogens 2022, 11, 344. https://doi.org/10.3390/pathogens11030344
Gallenti R, Hussein HE, Alzan HF, Suarez CE, Ueti M, Asurmendi S, Benitez D, Araujo FR, Rolls P, Sibeko-Matjila K, et al. Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies. Pathogens. 2022; 11(3):344. https://doi.org/10.3390/pathogens11030344
Chicago/Turabian StyleGallenti, Romina, Hala E. Hussein, Heba F. Alzan, Carlos E. Suarez, Massaro Ueti, Sebastián Asurmendi, Daniel Benitez, Flabio R. Araujo, Peter Rolls, Kgomotso Sibeko-Matjila, and et al. 2022. "Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies" Pathogens 11, no. 3: 344. https://doi.org/10.3390/pathogens11030344
APA StyleGallenti, R., Hussein, H. E., Alzan, H. F., Suarez, C. E., Ueti, M., Asurmendi, S., Benitez, D., Araujo, F. R., Rolls, P., Sibeko-Matjila, K., Schnittger, L., & Florin-Christensen, M. (2022). Unraveling the Complexity of the Rhomboid Serine Protease 4 Family of Babesia bovis Using Bioinformatics and Experimental Studies. Pathogens, 11(3), 344. https://doi.org/10.3390/pathogens11030344











