Development of a Mouse Model to Explore CD4 T Cell Specificity, Phenotype, and Recruitment to the Lung after Influenza B Infection
Abstract
1. Introduction
2. Results
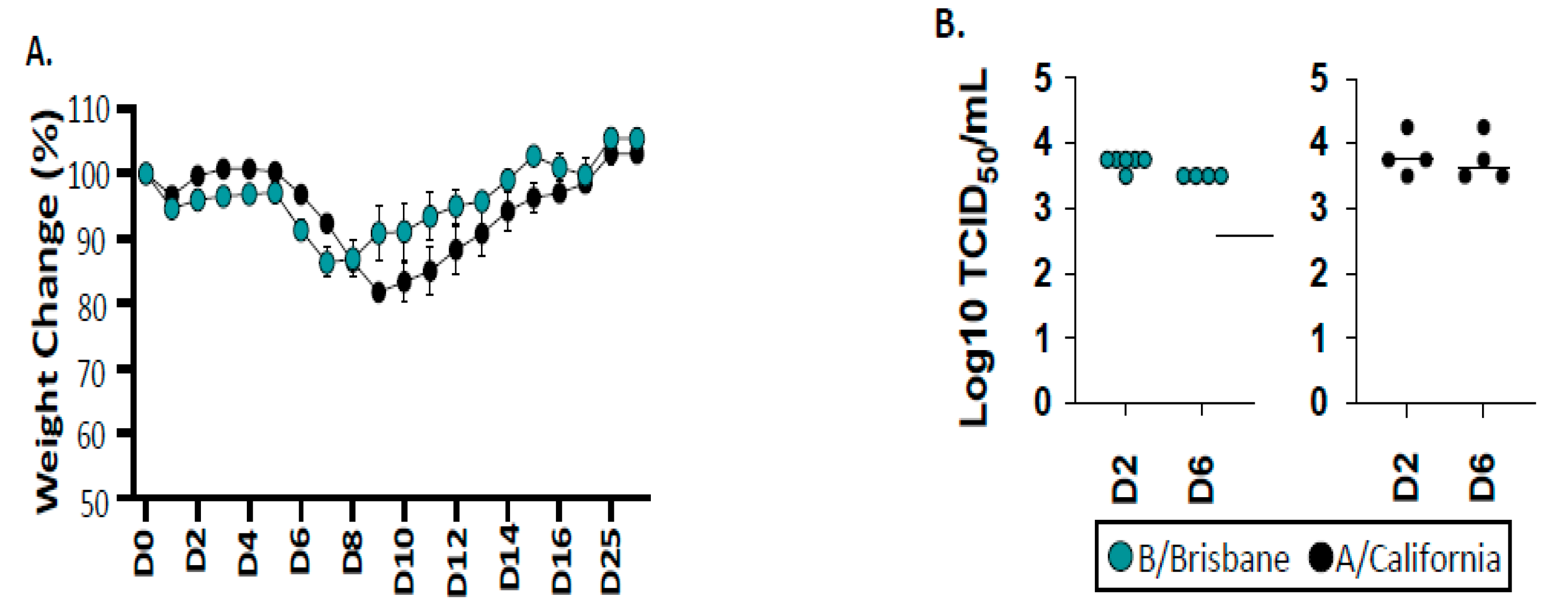
2.1. Infection and Viral Replication of Influenza B after Infection of B6 Mice
2.2. The Specificity, Abundance, and Localization of IBV-Specific CD4 T Cells
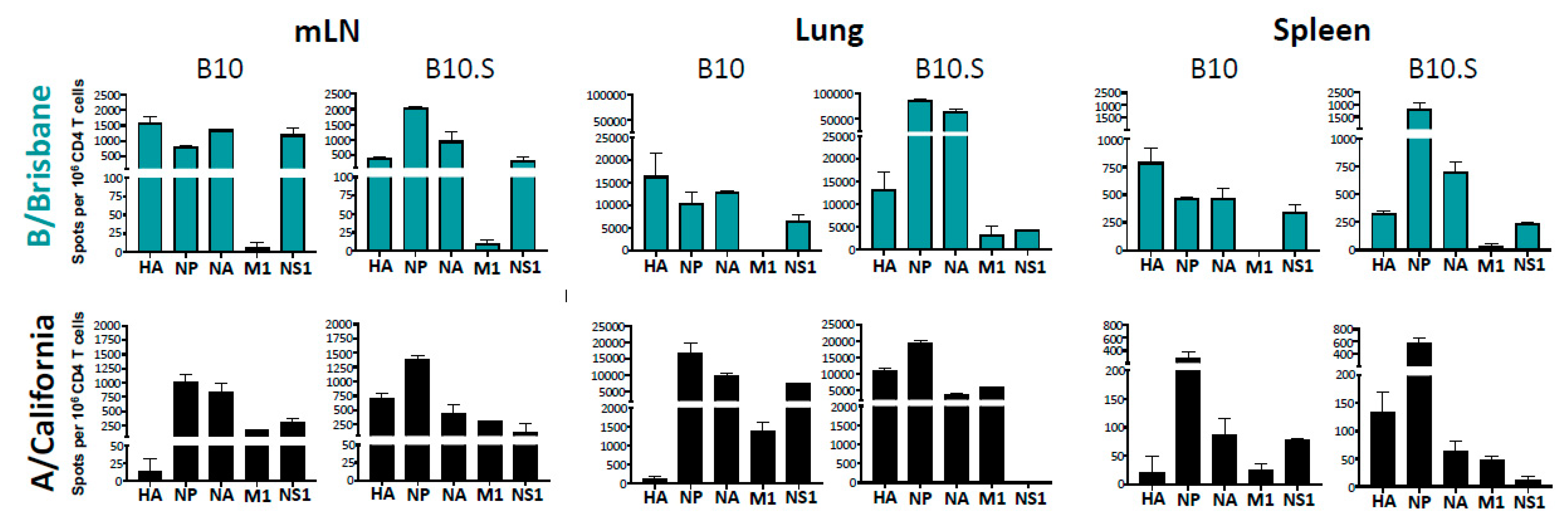
2.3. MHC-Dependent Viral Protein Biases in CD4 T Cell Specificity
2.4. Distribution of Cytokine-Producing IBV-Specific CD4 T Cells after Infection
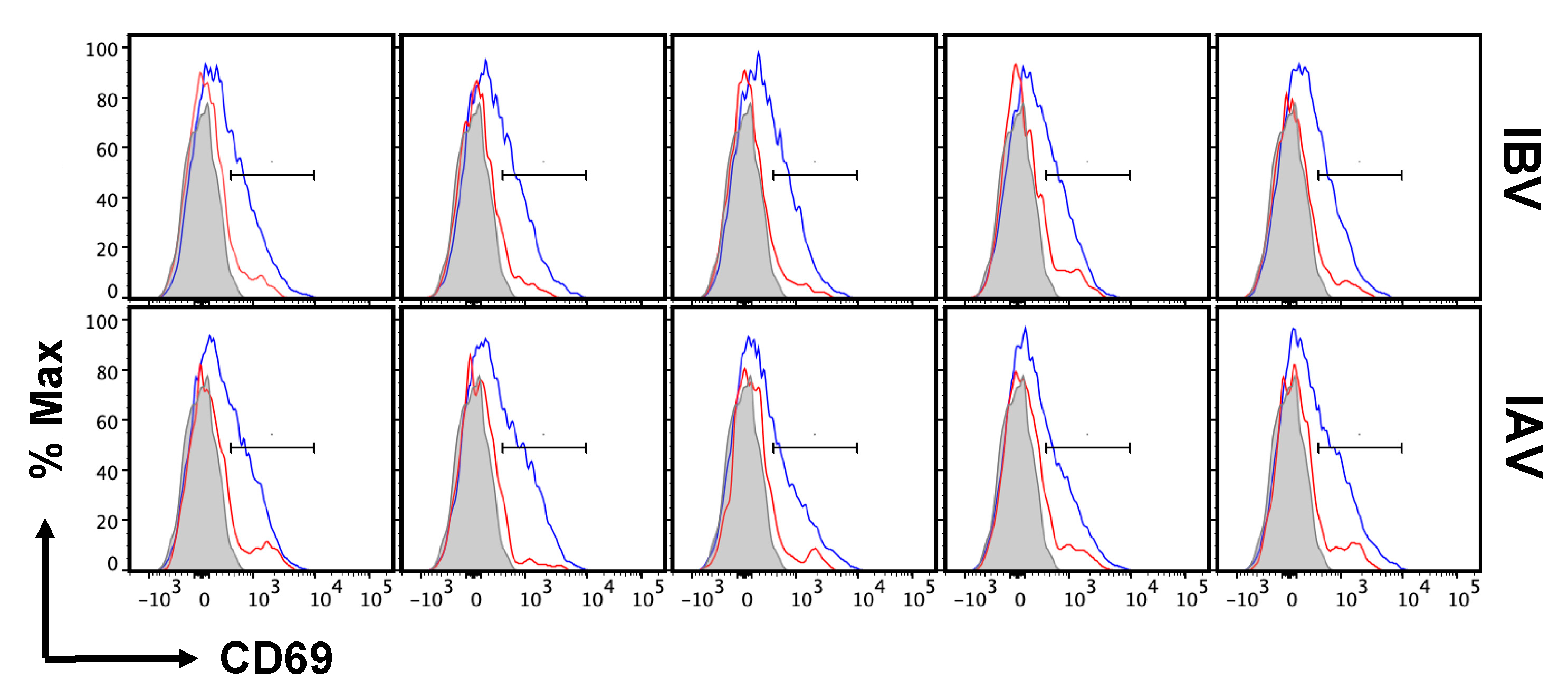
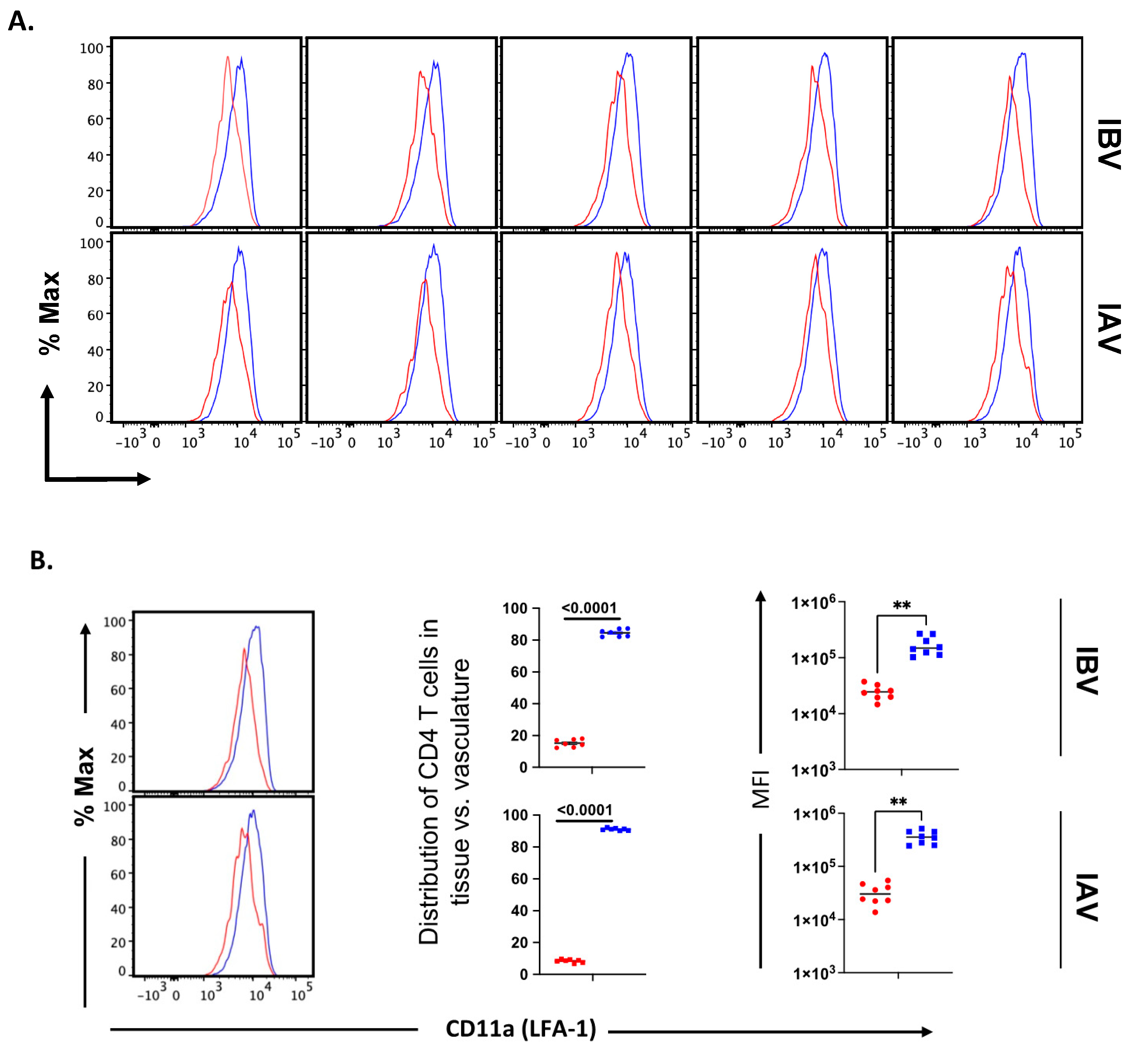
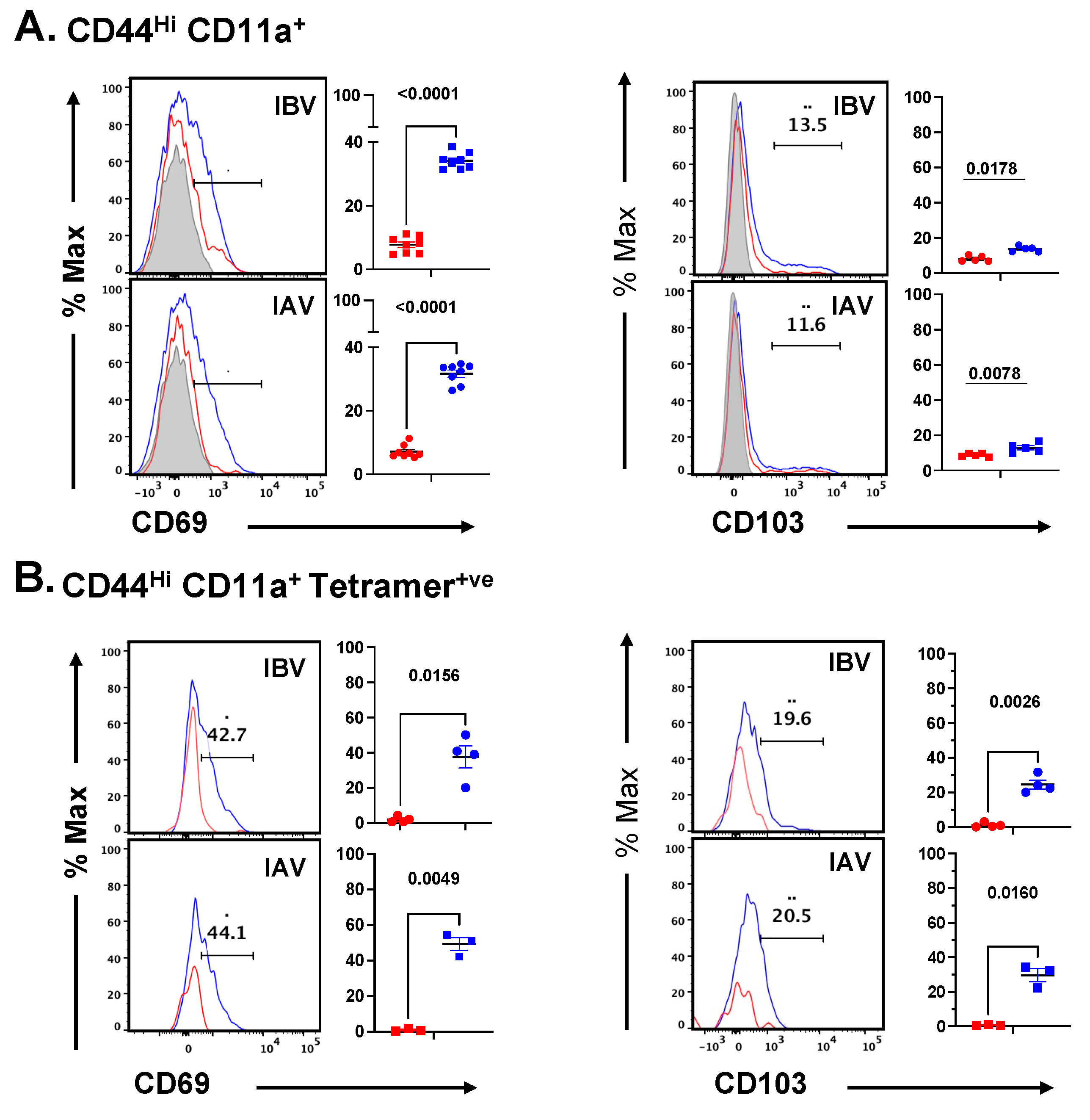
2.5. Distribution and Cell Surface Phenotype of IBV-Specific CD4 T Cells in the Lung
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Mice and Viruses
4.3. Peptides
4.4. Virus Infections
4.5. Lung Virus Titers and TCID50
4.6. EliSpot Assays
4.7. IV labeling, Tetramer Enrichment, and Flow Cytometry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Margine, I.; Krammer, F. Animal Models for Influenza Viruses: Implications for Universal Vaccine Development. Pathogens 2014, 3, 845–874. [Google Scholar] [CrossRef]
- Thangavel, R.R.; Bouvier, N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods 2014, 410, 60–79. [Google Scholar] [CrossRef]
- Tripp, R.A.; Tompkins, S.M. Animal Models for Evaluation of Influenza Vaccines. Curr. Top Microbiol. Immunol. 2009, 333, 397–412. [Google Scholar] [CrossRef]
- Bodewes, R.; Rimmelzwaan, G.F.; Osterhaus, A. Animal models for the preclinical evaluation of candidate influenza vaccines. Expert Rev. Vaccines 2010, 9, 59–72. [Google Scholar] [CrossRef]
- Roubidoux, E.; Schultz-Cherry, S. Animal Models Utilized for the Development of Influenza Virus Vaccines. Vaccines 2021, 9, 787. [Google Scholar] [CrossRef]
- Boon, A.C.M.; Finkelstein, D.; Zheng, M.; Liao, G.; Allard, J.; Klumpp, K.; Webster, R.; Peltz, G.; Webby, R.J. H5N1 Influenza Virus Pathogenesis in Genetically Diverse Mice Is Mediated at the Level of Viral Load. MBio 2011, 2, e00171-11. [Google Scholar] [CrossRef]
- Otte, A.; Sauter, M.; Alleva, L.; Baumgarte, S.; Klingel, K.; Gabriel, G. Differential Host Determinants Contribute to the Pathogenesis of 2009 Pandemic H1N1 and Human H5N1 Influenza A Viruses in Experimental Mouse Models. Am. J. Pathol. 2011, 179, 230–239. [Google Scholar] [CrossRef]
- Honce, R.; Schultz-Cherry, S. Impact of Obesity on Influenza A Virus Pathogenesis, Immune Response, and Evolution. Front. Immunol. 2019, 10, 1071. [Google Scholar] [CrossRef]
- Concannon, M.A.; Jiang, J. The effect of adjuvants on vaccine-induced antibody response against influenza in aged mice. Front. Biol. 2014, 9, 89–94. [Google Scholar] [CrossRef]
- Frasca, D.; Blomberg, B.B.; Garcia, D.; Keilich, S.R.; Haynes, L. Age-related factors that affect B cell responses to vaccination in mice and humans. Immunol. Rev. 2020, 296, 142–154. [Google Scholar] [CrossRef]
- Trammell, R.A.; Toth, L.A. Genetic susceptibility and resistance to influenza infection and disease in humans and mice. Expert Rev. Mol. Diagn. 2008, 8, 515–529. [Google Scholar] [CrossRef]
- Reneer, Z.B.; Jamieson, P.J.; Skarlupka, A.L.; Huang, Y.; Ross, T.M. Computationally Optimized Broadly Reactive H2 HA Influenza Vaccines Elicited Broadly Cross-Reactive Antibodies and Protected Mice from Viral Challenges. J. Virol. 2020, 95, e01526-20. [Google Scholar] [CrossRef]
- Boon, A.C.M.; Debeauchamp, J.; Hollmann, A.; Luke, J.; Kotb, M.; Rowe, S.; Finkelstein, D.; Neale, G.; Lu, L.; Williams, R.W.; et al. Host Genetic Variation Affects Resistance to Infection with a Highly Pathogenic H5N1 Influenza A Virus in Mice. J. Virol. 2009, 83, 10417–10426. [Google Scholar] [CrossRef]
- Srivastava, B.; Błażejewska, P.; Heßmann, M.; Bruder, D.; Geffers, R.; Mauel, S.; Gruber, A.D.; Schughart, K. Host Genetic Background Strongly Influences the Response to Influenza A Virus Infections. PLoS ONE 2009, 4, e4857. [Google Scholar] [CrossRef]
- Otte, A.; Gabriel, G. 2009 pandemic H1N1 influenza A virus strains display differential pathogenicity in C57BL/6J but not BALB/c mice. Virulence 2011, 2, 563–566. [Google Scholar] [CrossRef][Green Version]
- Noll, K.; Ferris, M.T.; Heise, M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe 2019, 25, 484–498. [Google Scholar] [CrossRef]
- Leist, S.R.; Baric, R.S. Giving the Genes a Shuffle: Using Natural Variation to Understand Host Genetic Contributions to Viral Infections. Trends Genet. 2018, 34, 777–789. [Google Scholar] [CrossRef]
- Mishina, M.; Sakimura, K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 2007, 58, 105–112. [Google Scholar] [CrossRef]
- Takeuchi, T.; Nomura, T.; Tsujita, M.; Suzuki, M.; Fuse, T.; Mori, H.; Mishina, M. Flp recombinase transgenic mice of C57BL/6 strain for conditional gene targeting. Biochem. Biophys. Res. Commun. 2002, 293, 953–957. [Google Scholar] [CrossRef]
- Yang, S.; Adaway, M.; Du, J.; Huang, S.; Sun, J.; Bidwell, J.P.; Zhou, B. NMP4 regulates the innate immune response to influenza A virus infection. Mucosal Immunol. 2020, 14, 209–218. [Google Scholar] [CrossRef]
- Guo, L.; Feng, K.; Wang, Y.C.; Mei, J.J.; Ning, R.T.; Zheng, H.W.; Wang, J.J.; Worthen, G.S.; Wang, X.; Song, J.; et al. Critical role of CXCL4 in the lung pathogenesis of influenza (H1N1) respiratory infection. Mucosal Immunol. 2017, 10, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Brincks, E.L.; Gurung, P.; Langlois, R.A.; Hemann, E.A.; Legge, K.L.; Griffith, T.S. The Magnitude of the T Cell Response to a Clinically Significant Dose of Influenza Virus Is Regulated by TRAIL. J. Immunol. 2011, 187, 4581–4588. [Google Scholar] [CrossRef] [PubMed]
- Krueger, P.D.; Goldberg, M.F.; Hong, S.-W.; Osum, K.C.; Langlois, R.A.; Kotov, D.I.; Dileepan, T.; Jenkins, M.K. Two sequential activation modules control the differentiation of protective T helper-1 (Th1) cells. Immunity 2021, 54, 687–701.e4. [Google Scholar] [CrossRef] [PubMed]
- Mayer-Barber, K.D.; Barber, D.L.; Shenderov, K.; White, S.D.; Wilson, M.S.; Cheever, A.; Kugler, D.; Hieny, S.; Caspar, P.; Nunez, G.; et al. Caspase-1 independent IL-1beta production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J. Immunol. 2010, 184, 3326–3330. [Google Scholar] [CrossRef] [PubMed]
- Fehlings, M.; Chakarov, S.; Simoni, Y.; Sivasankar, B.; Ginhoux, F.; Newell, E. Multiplex peptide-MHC tetramer staining using mass cytometry for deep analysis of the influenza-specific T-cell response in mice. J. Immunol. Methods 2018, 453, 30–36. [Google Scholar] [CrossRef]
- Ertelt, J.M.; Rowe, J.H.; Johanns, T.M.; Lai, J.C.; McLachlan, J.B.; Way, S.S. Selective Priming and Expansion of Antigen-Specific Foxp3−CD4+ T Cells during Listeria monocytogenes Infection. J. Immunol. 2009, 182, 3032–3038. [Google Scholar] [CrossRef]
- Tripathi, P.; Mitchell, T.C.; Finkelman, F.; Hildeman, D.A. Cutting Edge: Limiting Amounts of IL-7 Do Not Control Contraction of CD4+ T Cell Responses1. J. Immunol. 2007, 178, 4027–4031. [Google Scholar] [CrossRef]
- Kotov, D.I.; Jenkins, M.K. Peptide:MHCII Tetramer-Based Cell Enrichment for the Study of Epitope-Specific CD4+ T Cells. Curr. Protoc. Immunol. 2019, 125, e75. [Google Scholar] [CrossRef]
- Crowe, S.R.; Miller, S.C.; Woodland, D.L. Identification of protective and non-protective T cell epitopes in influenza. Vaccine 2006, 24, 452–456. [Google Scholar] [CrossRef]
- Wiley, J.A.; Hogan, R.J.; Woodland, D.L.; Harmsen, A.G. Antigen-specific CD8+ T cells persist in the upper respiratory tract following influenza virus infection. J. Immunol. 2001, 167, 3293–3299. [Google Scholar] [CrossRef]
- Valkenburg, S.A.; Gras, S.; Guillonneau, C.; La Gruta, N.; Thomas, P.G.; Purcell, A.; Rossjohn, J.; Doherty, P.C.; Turner, S.J.; Kedzierska, K. Protective Efficacy of Cross-Reactive CD8+ T Cells Recognising Mutant Viral Epitopes Depends on Peptide-MHC-I Structural Interactions and T Cell Activation Threshold. PLoS Pathog. 2010, 6, e1001039. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.G.; Brown, S.A.; Keating, R.; Yue, W.; Morris, M.Y.; So, J.; Webby, R.J.; Doherty, P.C. Hidden epitopes emerge in secondary influenza virus-specific CD8+ T cell responses. J. Immunol. 2007, 178, 3091–3098. [Google Scholar] [CrossRef]
- Duan, S.; Meliopoulos, V.A.; McClaren, J.L.; Guo, X.-Z.J.; Sanders, C.J.; Smallwood, H.; Webby, R.J.; Schultz-Cherry, S.L.; Doherty, P.C.; Thomas, P.G. Diverse Heterologous Primary Infections Radically Alter Immunodominance Hierarchies and Clinical Outcomes Following H7N9 Influenza Challenge in Mice. PLoS Pathog. 2015, 11, e1004642. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Guan, J.; Handel, A.; Tscharke, D.C.; Sidney, J.; Sette, A.; Wakim, L.M.; Sng, X.Y.X.; Thomas, P.G.; Croft, N.P.; et al. Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Kedzierska, K.; La Gruta, N.; Davenport, M.; Turner, S.J.; Doherty, P.C. Contribution of T cell receptor affinity to overall avidity for virus-specific CD8+ T cell responses. Proc. Natl. Acad. Sci. USA 2005, 102, 11432–11437. [Google Scholar] [CrossRef] [PubMed]
- Brincks, E.L.; Katewa, A.; Kucaba, T.A.; Griffith, T.S.; Legge, K.L. CD8 T cells utilize TRAIL to control influenza virus infection. J. Immunol. 2008, 181, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
- McMaster, S.R.; Wein, A.N.; Dunbar, P.R.; Hayward, S.L.; Cartwright, E.K.; Denning, T.L.; Kohlmeier, J.E. Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucosal. Immunol. 2018, 11, 1071–1078. [Google Scholar] [CrossRef]
- Hirst, G.K. Studies on the mechanism of adaptation of influenza virus to mice. J. Exp. Med. 1947, 86, 357–366. [Google Scholar] [CrossRef]
- Hensen, L.; Kedzierska, K.; Koutsakos, M. Innate and adaptive immunity toward influenza B viruses. Futur. Microbiol. 2020, 15, 1045–1058. [Google Scholar] [CrossRef]
- Koutsakos, M.; Nguyen, T.H.; Barclay, W.S.; Kedzierska, K. Knowns and unknowns of influenza B viruses. Futur. Microbiol. 2016, 11, 119–135. [Google Scholar] [CrossRef]
- Zaraket, H.; Hurt, A.C.; Clinch, B.; Barr, I.; Lee, N. Burden of influenza B virus infection and considerations for clinical management. Antivir. Res. 2020, 185, 104970. [Google Scholar] [CrossRef] [PubMed]
- van de Sandt, C.E.; Bodewes, R.; Rimmelzwaan, G.F.; de Vries, R.D. Influenza B viruses: Not to be discounted. Future Microbiol. 2015, 10, 1447–1465. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, J.-O.; Kim, J.-Y.; Jung, H.E.; Lee, H.K.; Chang, J. Single mucosal vaccination targeting nucleoprotein provides broad protection against two lineages of influenza B virus. Antivir. Res. 2019, 163, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Kim, E.-A.; Chang, J. A “Prime and Deploy” Strategy for Universal Influenza Vaccine Targeting Nucleoprotein Induces Lung-Resident Memory CD8 T cells. Immune Netw. 2021, 21, e28. [Google Scholar] [CrossRef]
- Treanor, J.; Wright, P.F. Immune correlates of protection against influenza in the human challenge model. Dev. Biol. 2003, 115, 97–104. [Google Scholar]
- de Jong, J.C.; Palache, A.M.; Beyer, W.E.; Rimmelzwaan, G.F.; Boon, A.C.; Osterhaus, A.D. Haemagglutination-inhibiting antibody to influenza virus. Dev. Biol. 2003, 115, 63–73. [Google Scholar]
- Rimmelzwaan, G.F.; McElhaney, J.E. Correlates of protection: Novel generations of influenza vaccines. Vaccine 2008, 26, D41–D44. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Hale, J.S.; Ahmed, R. Memory T Follicular Helper CD4 T Cells. Front. Immunol. 2015, 6, 16. [Google Scholar] [CrossRef]
- Vinuesa, C.G.; Linterman, M.A.; Yu, D.; MacLennan, I.C. Follicular Helper T Cells. Annu. Rev. Immunol. 2016, 34, 335–368. [Google Scholar] [CrossRef]
- Fazilleau, N.; Mark, L.; McHeyzer-Williams, L.J.; McHeyzer-Williams, M.G. Follicular Helper T Cells: Lineage and Location. Immunity 2009, 30, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef]
- Devarajan, P.; Jones, M.; Kugler-Umana, O.; Vong, A.M.; Xia, J.; Swain, S.L. Pathogen Recognition by CD4 Effectors Drives Key Effector and Most Memory Cell Generation Against Respiratory Virus. Front. Immunol. 2018, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Zens, K.D.; Farber, D.L. Memory CD4 T cells in influenza. Curr. Top. Microbiol. Immunol. 2015, 386, 399–421. [Google Scholar] [PubMed]
- Sant, A.J.; Richards, K.A.; Nayak, J. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr. Opin. Immunol. 2018, 53, 13–21. [Google Scholar] [CrossRef]
- Juno, J.A.; van Bockel, D.; Kent, S.J.; Kelleher, A.D.; Zaunders, J.J.; Munier, C.M. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front Immunol. 2017, 8, 19. [Google Scholar] [CrossRef]
- Devarajan, P.; Ebautista, B.; Vong, A.M.; McKinstry, K.K.; Strutt, T.M.; Swain, S.L. New Insights into the Generation of CD4 Memory May Shape Future Vaccine Strategies for Influenza. Front. Immunol. 2016, 7, 136. [Google Scholar] [CrossRef]
- Brown, D.M.; Lampe, A.T.; Workman, A.M. The Differentiation and Protective Function of Cytolytic CD4 T Cells in Influenza Infection. Front. Immunol. 2016, 7, 93. [Google Scholar] [CrossRef]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4+ T Cells, Their Differentiation and Function. Front Immunol. 2017, 8, 194. [Google Scholar] [CrossRef]
- Marathe, B.M.; Mostafa, H.H.; Vogel, P.; Pascua, P.N.Q.; Jones, J.C.; Russell, C.J.; Webby, R.J.; Govorkova, E.A. A pharmacologically immunosuppressed mouse model for assessing influenza B virus pathogenicity and oseltamivir treatment. Antivir. Res. 2017, 148, 20–31. [Google Scholar] [CrossRef]
- Richards, K.A.; Topham, D.; Chaves, F.A.; Sant, A.J. Cutting Edge: CD4 T Cells Generated from Encounter with Seasonal Influenza Viruses and Vaccines Have Broad Protein Specificity and Can Directly Recognize Naturally Generated Epitopes Derived from the Live Pandemic H1N1 Virus. J. Immunol. 2010, 185, 4998–5002. [Google Scholar] [CrossRef]
- Nelson, S.A.; Dileepan, T.; Rasley, A.; Jenkins, M.K.; Fischer, N.O.; Sant, A.J. Intranasal Nanoparticle Vaccination Elicits a Persistent, Polyfunctional CD4 T Cell Response in the Murine Lung Specific for a Highly Conserved Influenza Virus Antigen That Is Sufficient To Mediate Protection from Influenza Virus Challenge. J. Virol. 2021, 95, e0084121. [Google Scholar] [CrossRef] [PubMed]
- Tobery, T.W.; Wang, S.; Wang, X.-M.; Neeper, M.P.; Jansen, K.U.; McClements, W.L.; Caulfield, M.J. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J. Immunol. Methods 2001, 254, 59–66. [Google Scholar] [CrossRef]
- Richards, K.A.; Chaves, F.A.; Krafcik, F.R.; Topham, D.J.; Lazarski, C.A.; Sant, A.J. Direct Ex Vivo Analyses of HLA-DR1 Transgenic Mice Reveal an Exceptionally Broad Pattern of Immunodominance in the Primary HLA-DR1-Restricted CD4 T-Cell Response to Influenza Virus Hemagglutinin. J. Virol. 2007, 81, 7608–7619. [Google Scholar] [CrossRef] [PubMed]
- Nayak, J.L.; Richards, K.A.; Chaves, F.A.; Sant, A.J. Analyses of the Specificity of CD4 T Cells During the Primary Immune Response to Influenza Virus Reveals Dramatic MHC-Linked Asymmetries in Reactivity to Individual Viral Proteins. Viral Immunol. 2010, 23, 169–180. [Google Scholar] [CrossRef]
- Richards, K.A.; Chaves, F.A.; Sant, A.J. Infection of HLA-DR1 Transgenic Mice with a Human Isolate of Influenza A Virus (H1N1) Primes a Diverse CD4 T-Cell Repertoire That Includes CD4 T Cells with Heterosubtypic Cross-Reactivity to Avian (H5N1) Influenza Virus. J. Virol. 2009, 83, 6566–6577. [Google Scholar] [CrossRef]
- Ni, F.; Kondrashkina, E.; Wang, Q. Structural basis for the divergent evolution of influenza B virus hemagglutinin. Virology 2013, 446, 112–122. [Google Scholar] [CrossRef]
- An, Y.; Parsons, L.M.; Jankowska, E.; Melnyk, D.; Joshi, M.; Cipollo, J.F. N-Glycosylation of Seasonal Influenza Vaccine Hemagglutinins: Implication for Potency Testing and Immune Processing. J. Virol. 2019, 93, e01693-18. [Google Scholar] [CrossRef]
- Hawgood, S.; Brown, C.; Edmondson, J.; Stumbaugh, A.; Allen, L.; Goerke, J.; Clark, H.; Poulain, F. Pulmonary Collectins Modulate Strain-Specific Influenza A Virus Infection and Host Responses. J. Virol. 2004, 78, 8565–8572. [Google Scholar] [CrossRef]
- Reading, P.C.; Morey, L.S.; Crouch, E.C.; Anders, E.M. Collectin-mediated antiviral host defense of the lung: Evidence from influenza virus infection of mice. J. Virol. 1997, 71, 8204–8212. [Google Scholar] [CrossRef]
- Watson, A.; Madsen, J.; Clark, H.W. SP-A and SP-D: Dual Functioning Immune Molecules with Antiviral and Immunomodulatory Properties. Front. Immunol. 2021, 11, 622598. [Google Scholar] [CrossRef] [PubMed]
- Sant, A.J.; DiPiazza, A.; Nayak, J.L.; Rattan, A.; Richards, K.A. CD4 T cells in protection from influenza virus: Viral antigen specificity and functional potential. Immunol. Rev. 2018, 284, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.G.; Mayerbarber, K.D.; Sung, H.; Beura, L.K.; James, B.R.; Taylor, J.J.; Qunaj, L.; Griffith, T.S.; Vezys, V.; Barber, D.L.; et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 2014, 9, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.G.; Sung, H.; Skon, C.N.; Lefrancois, L.; Deisinger, A.; Vezys, V.; Masopust, D. Cutting Edge: Intravascular Staining Redefines Lung CD8 T Cell Responses. J. Immunol. 2012, 189, 2702–2706. [Google Scholar] [CrossRef]
- Uddbäck, I.; Cartwright, E.K.; Schøller, A.S.; Wein, A.; Hayward, S.L.; Lobby, J.; Takamura, S.; Thomsen, A.R.; Kohlmeier, J.E.; Christensen, J.P. Long-term maintenance of lung resident memory T cells is mediated by persistent antigen. Mucosal Immunol. 2020, 14, 92–99. [Google Scholar] [CrossRef]
- Van Braeckel-Budimir, N.; Varga, S.M.; Badovinac, V.P.; Harty, J.T. Repeated Antigen Exposure Extends the Durability of Influenza-Specific Lung-Resident Memory CD8+ T Cells and Heterosubtypic Immunity. Cell Rep. 2018, 24, 3374–3382.e3. [Google Scholar] [CrossRef]
- Morabito, K.; Ruckwardt, T.R.; Redwood, A.; Moin, S.; Price, D.; Graham, B.S. Intranasal administration of RSV antigen-expressing MCMV elicits robust tissue-resident effector and effector memory CD8+ T cells in the lung. Mucosal Immunol. 2016, 10, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Knudson, C.J.; Weiss, K.A.; Hartwig, S.M.; Varga, S.M. The Pulmonary Localization of Virus-Specific T Lymphocytes Is Governed by the Tissue Tropism of Infection. J. Virol. 2014, 88, 9010–9016. [Google Scholar] [CrossRef]
- DiPiazza, A.; Laniewski, N.; Rattan, A.; Topham, D.J.; Miller, J.; Sant, A.J. CD4 T Cell Epitope Specificity and Cytokine Potential Are Preserved as Cells Transition from the Lung Vasculature to Lung Tissue following Influenza Virus Infection. J. Virol. 2018, 92, e00377-18. [Google Scholar] [CrossRef]
- Richards, K.A.; DiPiazza, A.T.; Rattan, A.; Knowlden, Z.A.G.; Yang, H.; Sant, A.J. Diverse Epitope Specificity, Immunodominance Hierarchy, and Functional Avidity of Effector CD4 T Cells Established During Priming Is Maintained in Lung After Influenza A Virus Infection. Front. Immunol. 2018, 9, 655. [Google Scholar] [CrossRef]
- Turner, D.; Bickham, K.L.; Thome, J.J.T.; Kim, C.Y.; D’Ovidio, F.; Wherry, E.J.; Farber, D. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2013, 7, 501–510. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Turner, D.; Pham, Q.; Wherry, E.J.; Lefrancois, L.; Farber, D.L. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 2011, 187, 5510–5514. [Google Scholar] [CrossRef]
- Chu, K.; Batista, N.V.; Girard, M.; Law, J.C.; Watts, T.H. GITR differentially affects lung effector T cell subpopulations during influenza virus infection. J. Leukoc. Biol. 2020, 107, 953–970. [Google Scholar] [CrossRef]
- Strutt, T.M.; Dhume, K.; Finn, C.M.; Hwang, J.H.; Castonguay, C.; Swain, S.L.; McKinstry, K.K. IL-15 supports the generation of protective lung-resident memory CD4 T cells. Mucosal Immunol. 2017, 11, 668–680. [Google Scholar] [CrossRef]
- Marshall, N.B.; Vong, A.M.; Devarajan, P.; Brauner, M.D.; Kuang, Y.; Nayar, R.; Schutten, E.A.; Castonguay, C.H.; Berg, L.J.; Nutt, S.L.; et al. NKG2C/E Marks the Unique Cytotoxic CD4 T Cell Subset, ThCTL, Generated by Influenza Infection. J. Immunol. 2016, 198, 1142–1155. [Google Scholar] [CrossRef]
- Takamura, S.; Yagi, H.; Hakata, Y.; Motozono, C.; McMaster, S.R.; Masumoto, T.; Fujisawa, M.; Chikaishi, T.; Komeda, J.; Itoh, J.; et al. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 2016, 213, 3057–3073. [Google Scholar] [CrossRef]
- Suarez-Ramirez, J.E.; Chandiran, K.; Brocke, S.; Cauley, L.S. Immunity to Respiratory Infection Is Reinforced Through Early Proliferation of Lymphoid TRM Cells and Prompt Arrival of Effector CD8 T Cells in the Lungs. Front. Immunol. 2019, 10, 1370. [Google Scholar] [CrossRef]
- Shiow, L.R.; Rosen, D.B.; Brdickova, N.; Xu, Y.; An, J.; Lanier, L.L.; Cyster, J.G.; Matloubian, M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 2006, 440, 540–544. [Google Scholar] [CrossRef]
- Cibrián, D.; Sánchez-Madrid, F. CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol. 2017, 47, 946–953. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Masopust, D. Tissue-resident memory T cells. Immunity 2014, 41, 886–897. [Google Scholar] [CrossRef]
- Christophersen, A. Peptide-MHC class I and class II tetramers: From flow to mass cytometry. HLA 2019, 95, 169–178. [Google Scholar] [CrossRef]
- Nepom, G.T. MHC Class II Tetramers. J. Immunol. 2012, 188, 2477–2482. [Google Scholar] [CrossRef]
- Reijonen, H.; Kwok, W.W. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods 2003, 29, 282–288. [Google Scholar] [CrossRef]
- Taylor, J.J.; Jenkins, M.K. CD4+ memory T cell survival. Curr. Opin. Immunol. 2011, 23, 319–323. [Google Scholar] [CrossRef]
- Dileepan, T.; Malhotra, D.; Kotov, D.I.; Kolawole, E.M.; Krueger, P.D.; Evavold, B.D.; Jenkins, M.K. MHC class II tetramers engineered for enhanced binding to CD4 improve detection of antigen-specific T cells. Nat. Biotechnol. 2021, 39, 943–948. [Google Scholar] [CrossRef]
- Vollers, S.S.; Stern, L.J. Class II major histocompatibility complex tetramer staining: Progress, problems, and prospects. Immunology 2008, 123, 305–313. [Google Scholar] [CrossRef]
- Martinez, R.J.; Evavold, B.D. Lower Affinity T Cells are Critical Components and Active Participants of the Immune Response. Front. Immunol. 2015, 6, 468. [Google Scholar] [CrossRef]
- Tan, J.; Asthagiri Arunkumar, G.; Krammer, F. Universal influenza virus vaccines and therapeutics: Where do we stand with influenza B virus? Curr. Opin. Immunol. 2018, 53, 45–50. [Google Scholar] [CrossRef]
- Schotsaert, M.; García-Sastre, A. Inactivated influenza virus vaccines: The future of TIV and QIV. Curr. Opin. Virol. 2017, 23, 102–106. [Google Scholar] [CrossRef]
- Ray, R.; Dos Santos, G.; Buck, P.O.; Claeys, C.; Matias, G.; Innis, B.L.; Bekkat-Berkani, R. A review of the value of quadrivalent influenza vaccines and their potential contribution to influenza control. Hum. Vaccines Immunother. 2017, 13, 1640–1652. [Google Scholar] [CrossRef]
- Santos, J.J.S.; Finch, C.; Sutton, T.; Obadan, A.; Aguirre, I.; Wan, Z.; Lopez, D.; Geiger, G.; Gonzalez-Reiche, A.S.; Ferreri, L.; et al. Development of an Alternative Modified Live Influenza B Virus Vaccine. J. Virol. 2017, 91, e00056-17. [Google Scholar] [CrossRef]
- Nogales, A.; Perez, D.R.; Santos, J.; Finch, C.; Martínez-Sobrido, L. Reverse Genetics of Influenza B Viruses. Methods Mol. Biol. 2017, 1602, 205–238. [Google Scholar] [CrossRef]
- White, C.; Chiem, K.; Perez, D.; Santos, J.; Garcia, S.C.; Nogales, A.; Martínez-Sobrido, L. A New Master Donor Virus for the Development of Live-Attenuated Influenza B Virus Vaccines. Viruses 2021, 13, 1278. [Google Scholar] [CrossRef]
- Park, J.-G.; Ávila-Pérez, G.; Nogales, A.; Blanco-Lobo, P.; de la Torre, J.C.; Martínez-Sobrido, L. Identification and Characterization of Novel Compounds with Broad-Spectrum Antiviral Activity against Influenza A and B Viruses. J. Virol. 2020, 94, e02149-19. [Google Scholar] [CrossRef]
- Pascua, P.N.Q.; Marathe, B.M.; Bisen, S.; Webby, R.J.; Govorkova, E.A. Influenza B viruses from different genetic backgrounds are variably impaired by neuraminidase inhibitor resistance–associated substitutions. Antivir. Res. 2019, 173, 104669. [Google Scholar] [CrossRef]
- Burnham, A.; Baranovich, T.; Govorkova, E.A. Neuraminidase inhibitors for influenza B virus infection: Efficacy and resistance. Antivir. Res. 2013, 100, 520–534. [Google Scholar] [CrossRef]
- Prokopyeva, E.; Kurskaya, O.; Sobolev, I.; Solomatina, M.; Murashkina, T.; Suvorova, A.; Alekseev, A.; Danilenko, D.; Komissarov, A.; Fadeev, A.; et al. Experimental Infection Using Mouse-Adapted Influenza B Virus in a Mouse Model. Viruses 2020, 12, 470. [Google Scholar] [CrossRef]
- Oh, D.Y.; Panozzo, J.; Vitesnik, S.; Farrukee, R.; Piedrafita, D.; Mosse, J.; Hurt, A.C. Selection of multi-drug resistant influenza A and B viruses under zanamivir pressure and their replication fitness in ferrets. Antivir. Ther. 2017, 23, 295–306. [Google Scholar] [CrossRef]
- Moser, M.J.; Hatta, Y.; Gabaglia, C.; Sanchez, A.; Dias, P.; Sarawar, S.; Kawaoka, Y.; Hatta, M.; Neumann, G.; Bilsel, P. Single-replication BM2SR vaccine provides sterilizing immunity and cross-lineage influenza B virus protection in mice. Vaccine 2019, 37, 4533–4542. [Google Scholar] [CrossRef]
- DiPiazza, A.; Richards, K.; Poulton, N.; Sant, A.J. Avian and Human Seasonal Influenza Hemagglutinin Proteins Elicit CD4 T Cell Responses That Are Comparable in Epitope Abundance and Diversity. Clin. Vaccine Immunol. 2017, 24, e00548-16. [Google Scholar] [CrossRef]
- Alexander, J.; Bilsel, P.; del Guercio, M.-F.; Stewart, S.; Marinkovic-Petrovic, A.; Southwood, S.; Crimi, C.; Vang, L.; Walker, L.; Ishioka, G.; et al. Universal influenza DNA vaccine encoding conserved CD4+ T cell epitopes protects against lethal viral challenge in HLA-DR transgenic mice. Vaccine 2010, 28, 664–672. [Google Scholar] [CrossRef]
- Moise, L.; Tassone, R.; Latimer, H.; Terry, F.; Levitz, L.; Haran, J.P.; Ross, T.M.; Boyle, C.M.; Martin, W.D.; De Groot, A.S. Immunization with cross-conserved H1N1 influenza CD4+ T-cell epitopes lowers viral burden in HLA DR3 transgenic mice. Hum. Vaccines Immunother. 2013, 9, 2060–2068. [Google Scholar] [CrossRef]
- Eickhoff, C.S.; Terry, F.E.; Peng, L.; Meza, K.A.; Sakala, I.G.; Van Aartsen, D.; Moise, L.; Martin, W.D.; Schriewer, J.; Buller, R.M.; et al. Highly conserved influenza T cell epitopes induce broadly protective immunity. Vaccine 2019, 37, 5371–5381. [Google Scholar] [CrossRef] [PubMed]
- Cameron, T.O.; Norris, P.J.; Patel, A.; Moulon, C.; Rosenberg, E.S.; Mellins, E.D.; Wedderburn, L.; Stern, L.J. Labeling antigen-specific CD4+ T cells with class II MHC oligomers. J. Immunol. Methods 2002, 268, 51–69. [Google Scholar] [CrossRef]
- Baker, S.; Guo, H.; Albrecht, R.A.; Garcia-Sastre, A.; Topham, D.J.; Martínez-Sobrido, L. Protection against Lethal Influenza with a Viral Mimic. J. Virol. 2013, 87, 8591–8605. [Google Scholar] [CrossRef]
- DiPiazza, A.; Nogales, A.; Poulton, N.; Wilson, P.C.; Martínez-Sobrido, L.; Sant, A.J. Pandemic 2009 H1N1 Influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef]
- Alymova, I.V.; Kodihalli, S.; Govorkova, E.A.; Fanget, B.; Gerdil, C.; Webster, R.G. Immunogenicity and protective efficacy in mice of influenza B virus vaccines grown in mammalian cells or embryonated chicken eggs. J. Virol. 1998, 72, 4472–4477. [Google Scholar] [CrossRef]
- Rattan, A.; Richards, K.A.; Knowlden, Z.A.G.; Sant, A.J. Protein Vaccination Directs the CD4+ T Cell Response toward Shared Protective Epitopes That Can Be Recalled after Influenza Virus Infection. J. Virol. 2019, 93, e00947-19. [Google Scholar] [CrossRef]
- Rattan, A.; Pawar, S.D.; Nawadkar, R.; Kulkarni, N.; Lal, G.; Mullick, J.; Sahu, A. Synergy between the classical and alternative pathways of complement is essential for conferring effective protection against the pandemic influenza A(H1N1) 2009 virus infection. PLoS Pathog. 2017, 13, e1006248. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Trop. Med. Hyg. 1938, 27, 493–497. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rattan, A.; White, C.L.; Nelson, S.; Eismann, M.; Padilla-Quirarte, H.; Glover, M.A.; Dileepan, T.; Marathe, B.M.; Govorkova, E.A.; Webby, R.J.; et al. Development of a Mouse Model to Explore CD4 T Cell Specificity, Phenotype, and Recruitment to the Lung after Influenza B Infection. Pathogens 2022, 11, 251. https://doi.org/10.3390/pathogens11020251
Rattan A, White CL, Nelson S, Eismann M, Padilla-Quirarte H, Glover MA, Dileepan T, Marathe BM, Govorkova EA, Webby RJ, et al. Development of a Mouse Model to Explore CD4 T Cell Specificity, Phenotype, and Recruitment to the Lung after Influenza B Infection. Pathogens. 2022; 11(2):251. https://doi.org/10.3390/pathogens11020251
Chicago/Turabian StyleRattan, Ajitanuj, Chantelle L. White, Sean Nelson, Max Eismann, Herbey Padilla-Quirarte, Maryah A. Glover, Thamotharampillai Dileepan, Bindumadhav M. Marathe, Elena A. Govorkova, Richard J. Webby, and et al. 2022. "Development of a Mouse Model to Explore CD4 T Cell Specificity, Phenotype, and Recruitment to the Lung after Influenza B Infection" Pathogens 11, no. 2: 251. https://doi.org/10.3390/pathogens11020251
APA StyleRattan, A., White, C. L., Nelson, S., Eismann, M., Padilla-Quirarte, H., Glover, M. A., Dileepan, T., Marathe, B. M., Govorkova, E. A., Webby, R. J., Richards, K. A., & Sant, A. J. (2022). Development of a Mouse Model to Explore CD4 T Cell Specificity, Phenotype, and Recruitment to the Lung after Influenza B Infection. Pathogens, 11(2), 251. https://doi.org/10.3390/pathogens11020251





